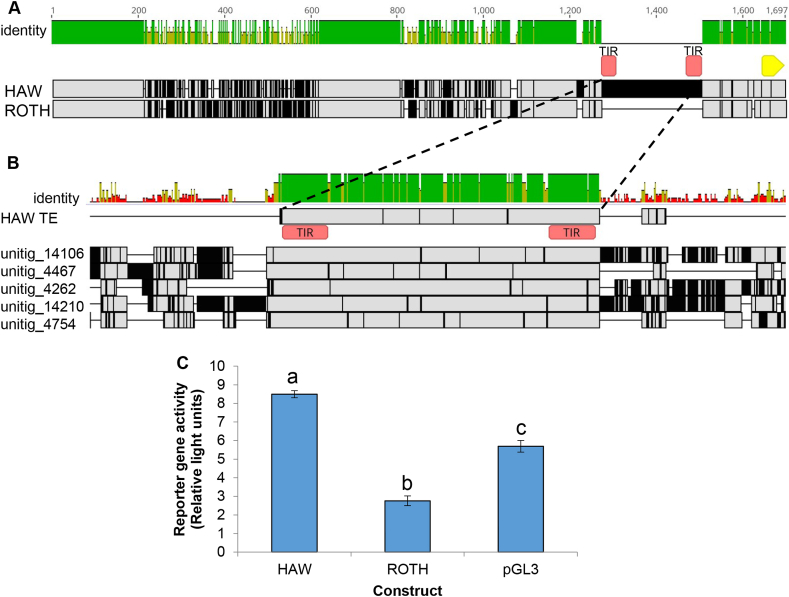
Fig. 4.
Alterations in the PxFMO2 promoter of the HAW strain enhance expression. A) Alignment of the sequences immediately upstream of PxFMO2 derived from the HAW and ROTH strains. The terminal inverted repeats (TIR) present at the boundaries of a putative transposable element insertion are indicated in red. The start of the coding sequence of PxFMO2 is shown in yellow. B) Alignment of the PxFMO2 genome against 5 representative scaffolds taken from the P. xylostella genome that share the repetitive element. In both A and B grey regions indicate similarity between sequences and black regions indicate sequence differences. Indels are represented by gaps in the sequence. The identity plot above the alignment displays the identity across all sequences for every position. Green indicates that the residue at the position is the same in all sequences. Yellow indicates less than complete identity and red refers to very low identity for the given position. C) Reporter gene activity (normalised to renilla fluorescence) of the ROTH and HAW PxFMO2 promoter variants. Letters above bars indicate significant differences, P < 0.001; one-way ANOVA with post-hoc Tukey HSD. Error bars indicate 95% CLs. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)