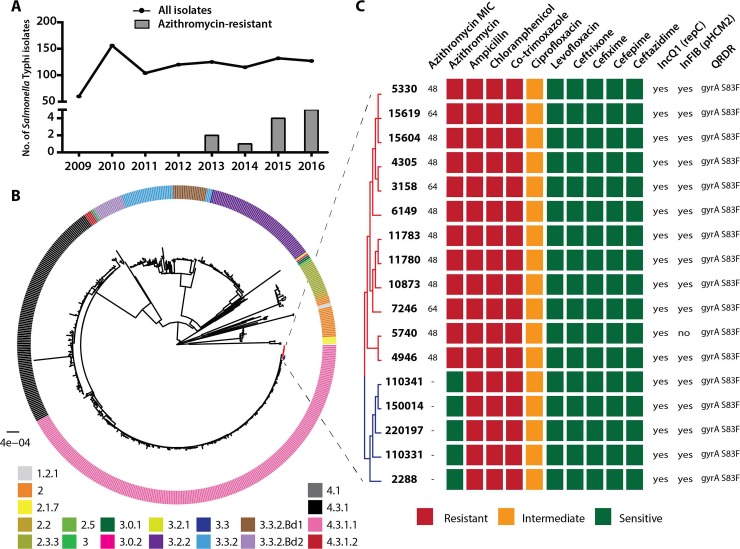
Fig 1. Emergence of azithromycin-resistant strains of Salmonella Typhi in Bangladesh and their genomic analysis.
(A) Temporal distribution of 939 Salmonella Typhi isolates included in the study. The number of isolates is shown as the line plot from 2009–2016. The numbers of azithromycin-resistant strains isolated each year is shown in the bar plot. Azithromycin-resistant strains were first isolated in 2013. (B) Whole-genome SNP tree of 548 Salmonella Typhi strains isolated in Bangladesh including 12 strains from the present study and 536 strains from a previous study [28]. The tree highlights the different genotypes that are found in Bangladesh including the most prevalent genotype 4.3.1.1 (H58 lineage 1). The 12 azithromycin-resistant strains (colored in red) clustered together within the genotype 4.3.1.1. Salmonella Typhimurium strain LT2 was used as an outgroup, while Salmonella Typhi strain CT18 was used as the reference strain. (C) Predicted and experimentally determined antimicrobial susceptibility pattern of azithromycin-resistant Salmonella Typhi strains and the most-closely related five azithromycin-sensitive strains. The antimicrobial susceptibility was experimentally determined through disc diffusion assay against a panel of 10 antibiotics. The predicted transmissible elements and antimicrobial resistance markers are also shown.