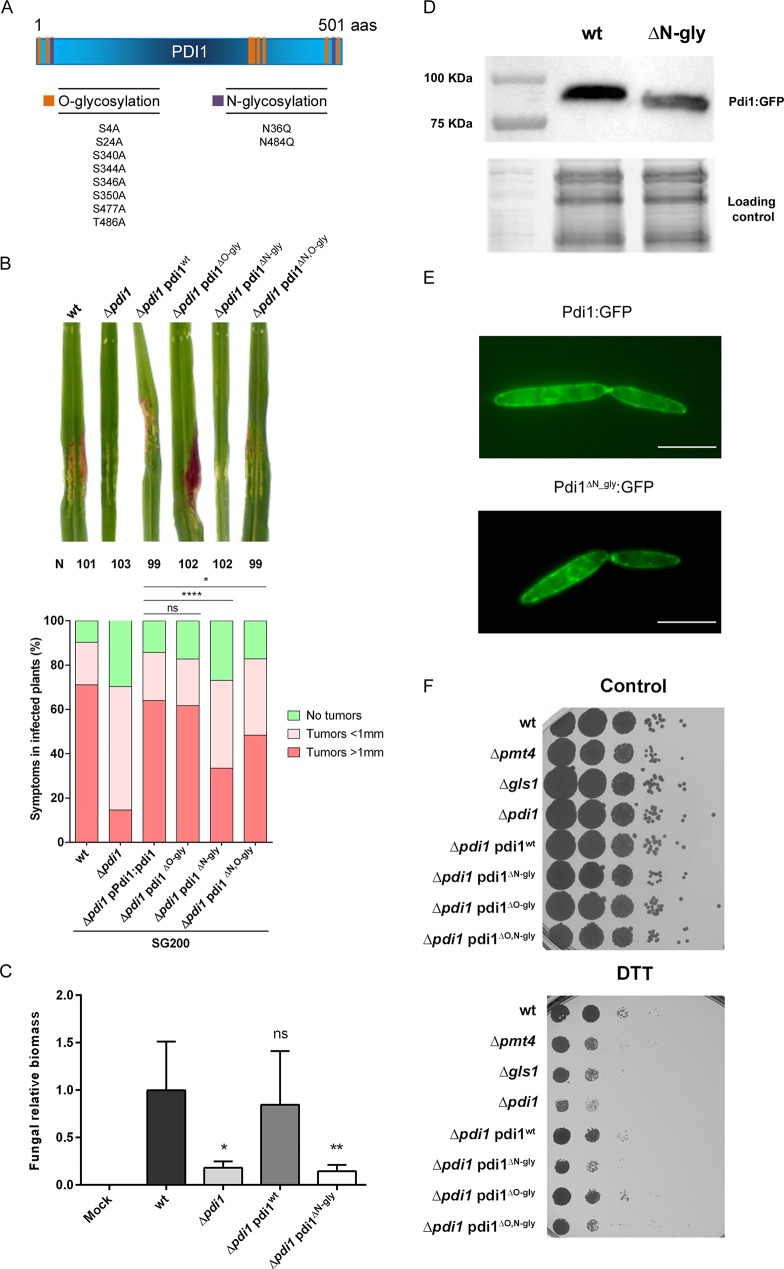
Fig 7. N-glycosylation of the protein disulfide isomerase Pdi1 ensures U. maydis virulence.
(A) Pdi1 has two putative N-glycosylation sites and eight putative O-glycosylation sites. The main amino acids where the glycosylation tree is anchored, serine/threonine in O-glycosylation and asparagine in N-glycosylation, have been replaced by similar amino acids alanine and glutamine for O- and N-glycosylation, respectively. (B) The percentage of symptoms in maize plants infected with the indicated strains at 14 dpi. Three independent infections with around 30 plants each were performed, and the total number of infected plants is indicated above each column. The Mann-Whitney statistical test was performed pooling the data for the three independent experiments (ns: not statistically significant; * for p-value < 0.05, **** for p-value < 0.001) or individually for each experiment as shown in S4 Table. Representative disease symptoms are shown above. (C) Quantification of fungal biomass in planta at 5 dpi was performed by qPCR, measuring the constitutively-expressed ppi1 U. maydis gene normalized to the constitutively-expressed plant gapdh gene. T-test statistical analysis was performed (ns for not statistically significant, * for p-value < 0.05, ** for p-value < 0.01). (D) Western blot showing Pdi1:GFP in SG200 and Pdi1ΔN-gly: GFP in SG200 Δpdi1, with the image of the stain-free gel activation as a loading control. (E) ER location for Pdi1:GFP and Pdi1ΔN-gly:GFP. Scale bar represents 10 μm. (F) Indicated strains were spotted onto CM plates supplemented with 2% D-glucose and 4 mM DTT. The mutation of N-glycosylation sites resulted on high DTT sensitivity.