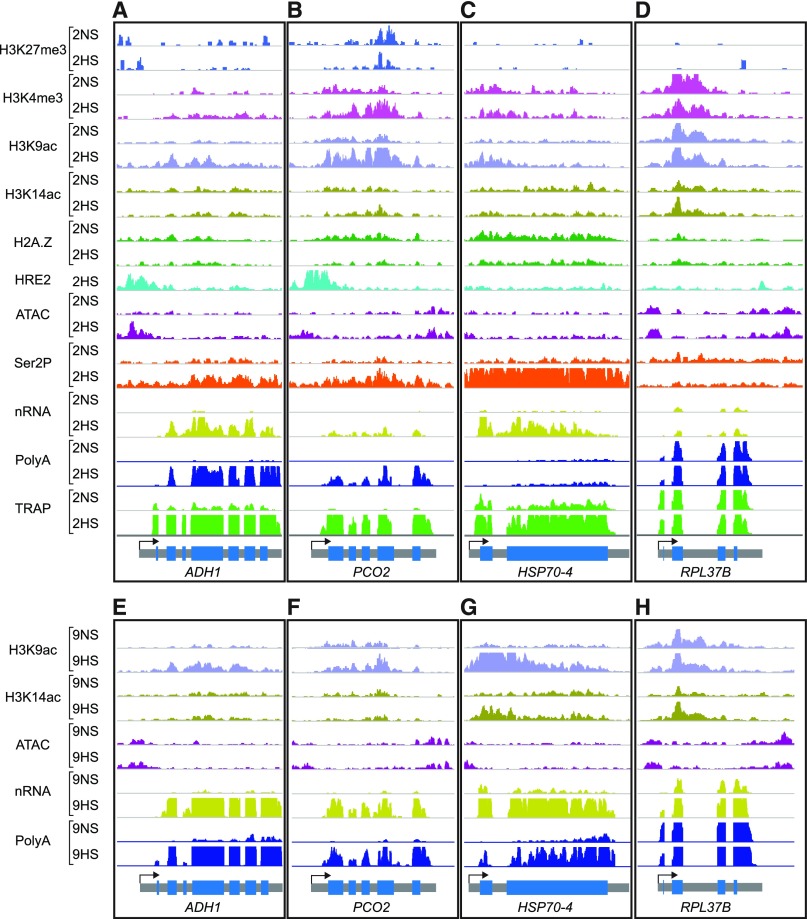
Figure 5.
Genome Browser View of Normalized Read Coverage of Histone, ATAC-seq, RNAPII–Ser2P, HRE2-ChIP, and RNA Outputs for Representative Genes
(A) to (D) Samples under 2NS and 2HS conditions.
(E) to (H) Samples under 9NS and 9HS conditions.
(A) and (E) ADH1 (Cluster 1), (B) and (F) PCO2 (Cluster 1), (C) and (G) HSP70-4 (Cluster 2), and (D) and (H) RPL37B (Cluster 9). The maximum read scale value used for chromatin-based and RNA-based outputs is equivalent for all genes depicted. At the bottom, the transcription unit is shown in gray with the TSS marked with an arrow. PCO2 shows a stronger decline in H2A.Z than ADH1 in response to hypoxia at 2HS (PCO2, −1.15 log2 FC; ADH1, −0.33 log2 FC) and provides an example of elevated H3K4me3 levels under HS.