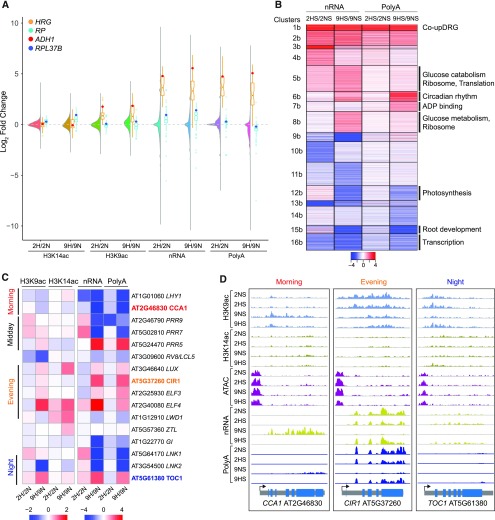
Figure 6.
Prolonged Hypoxia Leads to Further Variation in Epigenetic, Transcriptional, and Posttranscriptional Regulation.
(A) Combined violin and boxplot of H3K9ac and H3K14ac, nRNA, and polyA RNA data. Violin plots of log2 FC (9HS/9NS) data for all genes. Boxplots are data for the 49 HRGs plotted in orange with ADH1 depicted as a red dot and data for the cytosolic RPs plotted in blue, with RP37B depicted as a dark blue dot.
(B) Heatmap showing similarly regulated genes based on two dynamics in nRNA and polyA RNA. Partitioning around medoids clustering of 3,897 differentially regulated genes; 16 clusters. Selected enriched GO terms are shown at right (P adjusted < 1.37E-06), |log2 FC| > 1, FDR < 0.05.
(C) Heatmap of chromatin and RNA readouts of select circadian-regulated genes under short and prolonged HS. Heatmap scales between chromatin and RNA-based readouts differ.
(D) Brower views of a morning (CCA1), evening (CIR1, CIRCADIAN1), and night (TOC1) expressed gene. RNA scale of TOC1 is threefold lower than the scale used for CCA1 and CIR1.