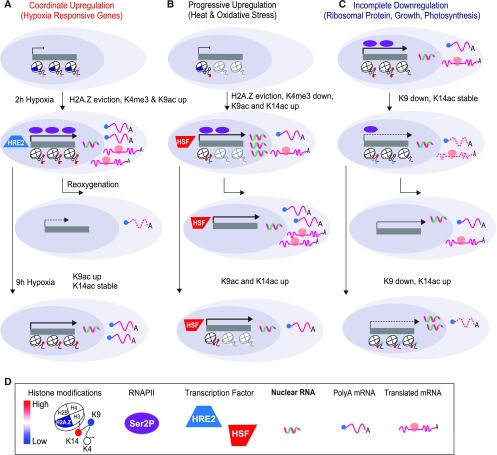
Figure 9.
Three Patterns of Dominant Nuclear Regulation in Response to Two Durations of HS and Reaeration
(A) Coordinate upregulation from chromatin to translation of HRGs characterized by hypoxia-triggered H2A.Z eviction and both H3K4me3 and H3K9ac elevation. Over half of the genes with this pattern of regulation showed HRE2 binding by ChIP. Transcript abundance and translation declines upon reoxygenation.
(B) Progressive upregulation of genes associated with heat and oxidative stress responses characterized by a strong 5′ bias in histone marks, hypoxia-triggered H2A.Z eviction, H3K9ac elevation, and H3K4me3 reduction. Many are known targets of HSFs. These genes are transcribed at low levels under normoxia (nonstress conditions), with evidence of transcriptional activation in response to brief hypoxia (2 h of hypoxia), as shown by elevated RNAPII–Ser2P binding, nRNA, and TRAP RNA. However, the elevation in polyA RNA was limited until the stress was prolonged (9 h hypoxia). Transcript abundance increases upon reoxygenation.
(C) Compartmentalized downregulation of many genes associated with growth and photosynthesis, including RP genes characterized by maintained RNAPII–Ser2P and nRNA levels under the stress, reduced H3K9ac, and progressive acquisition of H3K14ac. Transcript abundance and translation are restored upon reoxygenation. Cartoons illustrate histone modifications as defined by the key. HSF transcriptional activator; Ser2P, RNAPII with the Ser2P modification.
(D) Figure key for distinct outputs of gene regulation. Levels of assocation (high, red; low, blue) of Histone H3 modifications (H3K4me3, H3K9ac, H3K14ac) and the histone variant H2A.Z are depicted. Icons depicting binding by the transcription factors HRE2 and HSF, and RNAPII-Ser2P. Accumulation of nulcear, whole-cell, and ribosome associated RNA populations is also depicted.