Figure 1.
miR156-SPL9 Module Regulates Wax Synthesis.
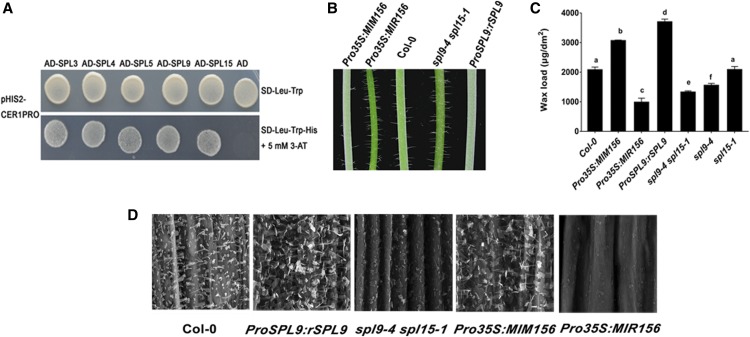
(A) Yeast one-hybrid assay to dissect the binding of SPLs to CER1 promoter DNA. For yeast one-hybrid experiment, CER1 promoter DNA region was fused to the HIS3 (auxotrophic marker) reporter gene in pHIS2 plasmid and tested for AD-SPLs binding. Empty pGADT7 vector was used as negative control.
(B) Glossy green and white waxy phenotypes of 6-week-old inflorescence stems of Pro35S:MIR156, Pro35S:MIM156, and SPL9 related lines compared with the wild type (Col-0). Pro35S:MIR156 and Pro35S:MIM156 are transgenic lines overexpressing miR156 and its artificial miRNA target mimic construct, respectively. ProSPL9:rSPL9 and spl9-4 spl15-1 represent SPL9 gain-of-function and loss-of-function plants, respectively.
(C) Cuticular wax amounts of inflorescence stems from 6-week-old wild-type, Pro35S:MIR156, Pro35S:MIM156, and SPL9 related Arabidopsis lines, which were grown under long-day conditions (16 h of light/8 h of dark). Cuticular waxes were extracted with hexane and analyzed by GC-FID. Wax coverage is expressed as wax amounts per stem surface area (μg.dm-2). Error bars indicate means from four replicate experiments, and their SD are shown. The significant differences between samples labeled with different lowercase letters were determined using ANOVA with a post hoc Tukey Honestly Significant Difference test.
(D) SEM images of epicuticular wax crystals on inflorescence stems of 6-week-old wild-type, Pro35S:MIR156, Pro35S:MIM156, and SPL9 related Arabidopsis lines. Bars = 5 μm.