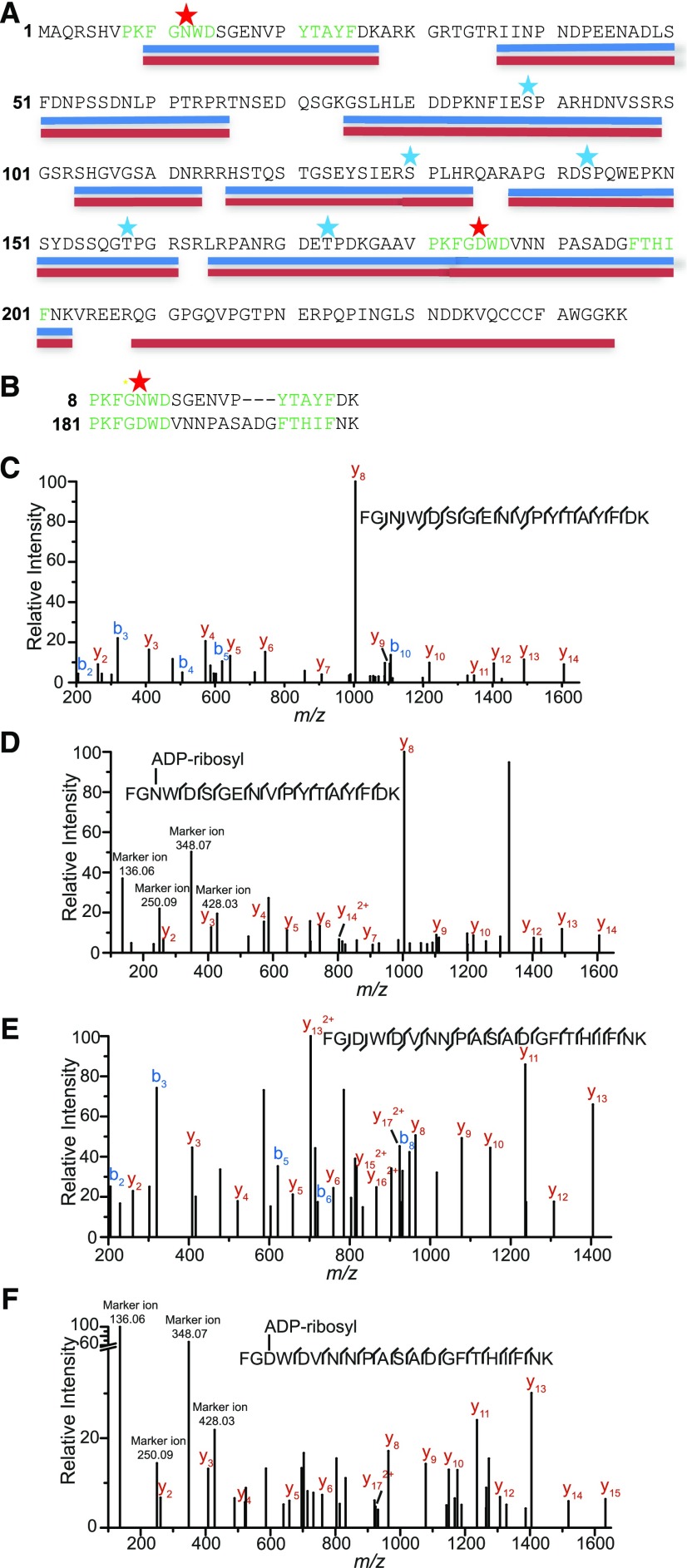
Figure 2.
GmRIN4b Is ADP Ribosylated in Vivo in the Presence of AvrRpm1.
sYFP-GmRIN4b was transiently expressed with AvrRpm1 or an EV control in N. benthamiana, and purified YFP-GmRIN4b protein was subjected to trypsin digestion prior to analysis by liquid chromatography–tandem mass spectrometry.
(A) GmRIN4b coverage map. Peptides detected in the negative control EV samples are underlined in blue. Peptides detected in the AvrRpm1-treated samples are underlined in red, with red stars indicating potentially ADP-ribosylated residues (only found in the AvrRpm1 samples; Supplemental Data Set 1). Blue stars indicate phosphorylated residues found in both EV and AvrRpm1-treated samples. The highly conserved PxFGxWD and F/YTxxFxK motifs of the NOI domains are highlighted in green.
(B) Alignment of the N- (top) and C-terminal (bottom) NOI domain peptides found to be modified by AvrRpm1. Star indicates position of ADP-ribosylation.
(C) Mass spectra resulting from fragmentation of the GmRIN4b N-terminal NOI domain peptide FGNWDSGENVPYTAYFDK, which was expressed in the absence of AvrRpm1.
(D) Mass spectra resulting from fragmentation of the GmRIN4b N-terminal NOI domain peptide FGNWDSGENVPYTAYFDK, which was expressed in the presence of AvrRpm1.
(E) Mass spectra resulting from fragmentation of the GmRIN4b C-terminal NOI domain peptide FGDWDVNNPASADGFTHIFNK, which was expressed in the absence of AvrRpm1.
(F) Mass spectra resulting from fragmentation of the GmRIN4b C-terminal NOI domain peptide FGDWDVNNPASADGFTHIFNK, which was expressed in the presence of AvrRpm1.
See also Supplemental Data Set 1.