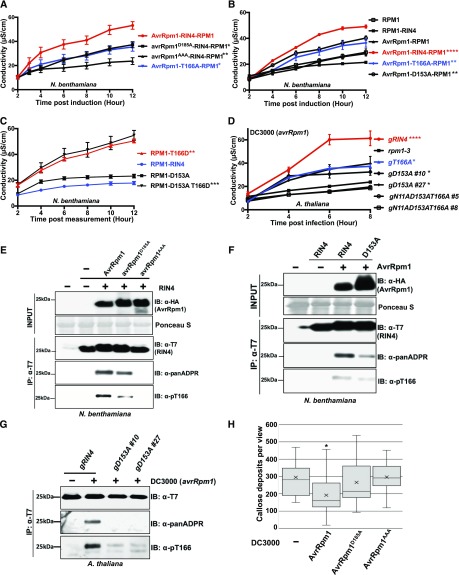
Figure 4.
ADP-Ribosylation of AtRIN4 by AvrRpm1 Is Required for AtRIN4 T166 Phosphorylation.
(A) Proposed AvrRpm1 ADP-ribosyl transferase catalytic residues are required for activation of RPM1 in N. benthamiana. The indicated AvrRpm1 mutants were coexpressed with genomic RPM1-myc and AtRIN4. The AtRIN4 T166A mutant was used as a control as it is known to partially suppress RPM1 activation. Cell death induced by RPM1 activation was monitored by conductivity measurements following induction of AvrRpm1 expression with 20 μM estradiol. Statistical analyses were performed by using one-way analysis of variance at 95% confidence comparing the values for the indicated AvrRpm1 and RIN4 mutant derivatives to the wild-type AvrRpm1-RIN4-RPM1 combination at 6 h postinduction. Asterisks represent statistical differences with the indicated P-values (****P < 0.0001, ***P < 0.005, **P < 0.001, *P < 0.05).
(B) D153 ribosylation site of AtRIN4 contributes to full AvrRpm1-mediated RPM1 activation in N. benthamiana. The wild-type AtRIN4, AtRIN4T166A (T166A), or AtRIN4D153A (D153A) were co-infiltrated with RPM1-myc and the wild-type AvrRpm1. Cell death was quantified by conductivity measurements as described in (A). Statistical analyses were performed as in (A), but using the wild-type RPM1-RIN4 in the absence of AvrRpm1 as the reference.
(C) Cell death mediated by the AtRIN4T166D (T166D) phosphomimic is epistatic to the loss of ribosylation at AtRIN4D153A (D153A). The quantification of cell death was monitored as in (A) and (B). Statistical analyses were performed as in (B).
(D) Ribosylation at AtRIN4D153 and phosphorylation of AtRIN4T166 contribute additively to AvrRpm1-mediated activation of RPM1 in transgenic Arabidopsis plants. P. syringae strain DC3000 (avrRpm1) was inoculated (5 × 107 cfu/mL) into leaves of transgenic Arabidopsis lines expressing genomic versions of AtRIN4 wild type (gRIN4), AtRIN4T166A (gT166A), AtRIN4D153A (gD153A), and AtRIN4N11AD153AT166A (gN11AD153AT166A) mutant plants. The rpm1-3 null mutant was used as a negative control. The quantification of cell death was monitored as in (A) and (B). Statistical analyses were performed as in (A), but using rpm1-3 as the reference.
(E) ADP-ribosylation activity of AvrRpm1 is required for full phosphorylation of AtRIN4T166. The ADP-ribosylation and phosphorylation of AtRIN4 in N. benthamiana were monitored by immunoprecipitation of AtRIN4 with anti-T7 followed by immunoblots with anti-T7, anti-panADPR, or anti-pT166. Co-infiltration of AvrRpm1 alleles and AtRIN4 was performed as in (A). Samples were collected 4 h posttreatment with 20 μM estradiol. This result represents one of four independent experiments. −, absence; +, presence; IB, immunoblot; IP, immunoprecipitation.
(F) ADP-ribosylation at AtRIN4D153 is required for full phosphorylation of Thr-166. N. benthamiana leaves were co-infiltrated with AvrRpm1 and AtRIN4 or AtRIN4D153A, as in (A). Samples were taken 12 h postinduction of AvrRpm1 with 20 μM estradiol. The ADP-ribosylation and phosphorylation of AtRIN4 in N. benthamiana were monitored by immunoprecipitation of AtRIN4 with anti-T7 followed by immunoblots with anti-T7, anti-panADPR, or anti-pT166. This result represents one of five independent experiments. −, absence; +, presence; IB, immunoblot; IP, immunoprecipitation.
(G) ADP-ribosylation at AtRIN4D153 is necessary for full phosphorylation of T166 in Arabidopsis. Transgenic plants expressing the wild-type AtRIN4 (gRIN4) and two independent T2 plants expressing AtRIN4D153A (gD153A #10 and #27) were inoculated with DC3000(avrRpm1) as in (D). Samples were collected 4 h postinfection before hypersensitive response developed. Immunoprecipitation of RIN4 proteins with anti-T7 was followed by immunoblots with anti-T7, anti-panADPR, and anti-pT166. −, absence; +, presence; IB, immunoblot; IP, immunoprecipitation.
(H) AvrRpm1 ADP-ribosylation catalytic residues are required for a virulence function of AvrRpm1 in Arabidopsis. Arabidopsis rpm1-3 mutant plants were inoculated with 5 × 107 cfu/mL of P. syringae strains DC3000 (EV), DC3000 (avrRpm1), DC3000 (avrRpm1D185A), or DC3000 (avrRpm1AAA). Callose accumulation from Arabidopsis leaves 18 h postinfection was analyzed by aniline blue staining. The box plot indicates mean (X), median (inner line), inner quartiles (box), outer quartiles (whiskers), and outliers (dots) for composite data from 16 samples (n = 16). Statistical significances compared with DC3000 (EV)-infected samples were determined by Student’s t test; *P < 0.05. This result was confirmed in three independent experiments. See also Supplemental Figure 2.