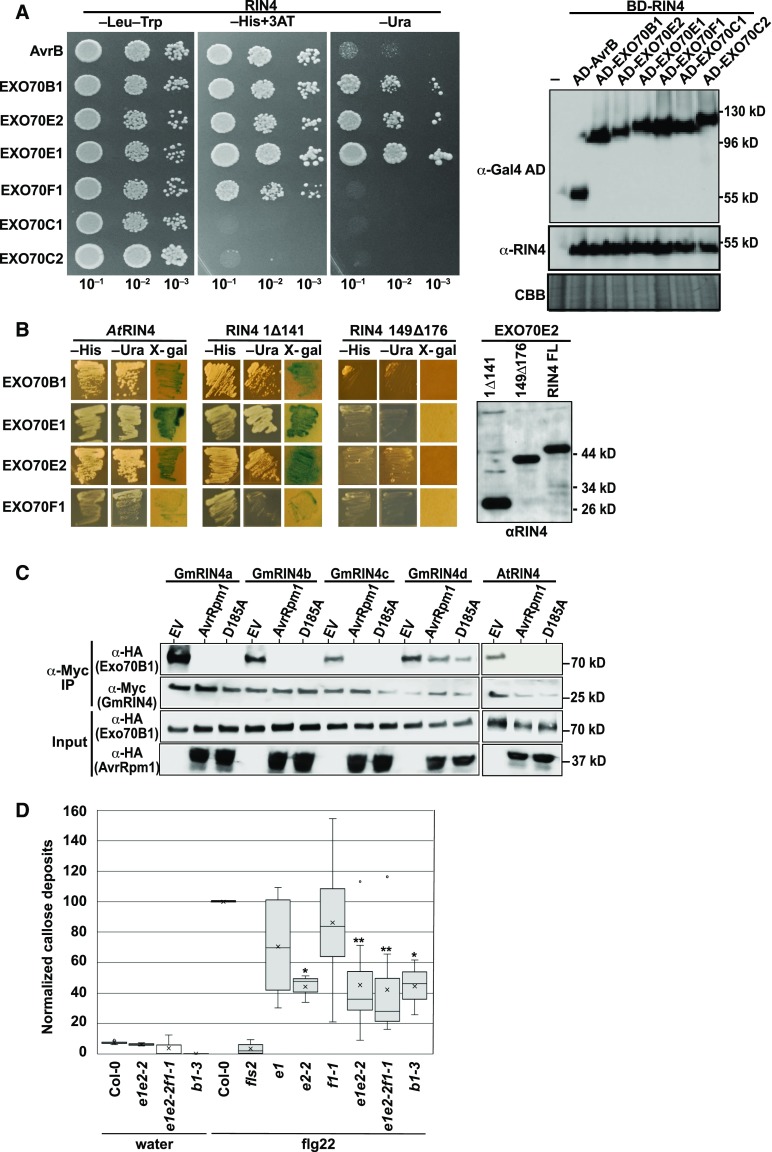
Figure 5.
RIN4 Proteins Interact with EXO70 Proteins via the C-NOI Domain.
(A) AtRIN4 interacts with multiple Arabidopsis EXO70 family members in yeast. Shown are the results of a yeast two-hybrid assay, with yeast expressing the indicated protein fusions. The AvrB protein was used as a positive control for interaction with AtRIN4. Growth on −His and −Ura plates indicate positive interactions. The immunoblot on the right confirms protein expression of each bait and prey construct. CBB, Coomassie Brilliant Blue.
(B) C-NOI domain of AtRIN4 is required for interaction with Arabidopsis EXO70 proteins. 1Δ141 indicates deletion of AtRIN4 amino acids 1 to 141, which contains the N-NOI domain. 149Δ176 indicates deletion of amino acids 149 to 176, which contains the C-NOI domain. The anti-RIN4 immunoblot shows that all three RIN4 protein fusions were expressed in yeast.
(C) AvrRpm1 inhibits association of Arabidopsis EXO70B1 with Arabidopsis and soybean RIN4 proteins. The indicated proteins were coexpressed in N. benthamiana, and then RIN4 proteins were immunoprecipitated with anti-Myc antibody. EXO70B1 coimmunoprecipitated in the absence of AvrRpm1, but not in its presence. D185A indicates AvrRpm1 with the D185A substitution. This result represents one of three independent experiments. IP, immunoprecipitation.
(D) Leaves from Arabidopsis plants were infiltrated with water or 100 μM flg22, and callose deposits were enumerated after 16 h. Shown is a box plot indicating mean (X), median (inner line), inner quartiles (box), outer quartiles (whiskers), and outliers (dots) for composite data from 13 independent experiments (for flg22 treatments, n = 13 [ecotype Columbia-0 (Col-0)], 6 [fls2, exo70e1, exo70 f1-1], 8 [exo70e1exo70e2-2 and exo70e1exo70e2-2exo70f1-1], and 3 [exo70e2-2 and exo70b1-3]), with the number of callose deposits detected in flg22-infiltrated Col-0 normalized to 100 within each experiment. Significance for flg22-treated mutant plants compared with flg22-treated Col-0 plants were determined by Student’s t test (*P < 0.05, **P < 0.005).
See also Supplemental Figures 3 and 4.