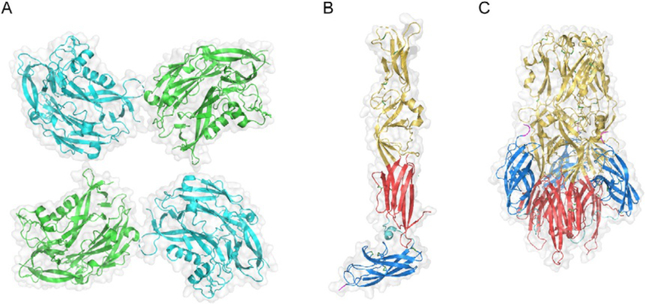
Fig. 3.
Structure of hantavirus Gn and Gc. (A) Partial structural model for Gn tetramer derived by fitting the X-ray crystal structure of the N-terminal region of PUUV Gn (amino acid residues 29–383; PDB ID: 5FXU) into the cryo-electron tomography (cryo-ET)-derived density map of the TULV glycoprotein spike (PDB ID: 5FYN) (Li et al., 2016). Adjacent subunits are colored in cyan and green. Gray outlines in (A)–(C) represent surface-shaded views. En face view relative to the surface of the viral particle is shown. (B) X-ray crystal structure of the monomeric ectodomain of HTNV Gc in its putative pre-fusion conformation (PDB ID: 5LJY) (Guardado-Calvo et al., 2016). (C) X-ray crystal structure of the ectodomain of HTNV Gc in its trimeric post-fusion conformation (PDB ID: 5LK0) (Guardado-Calvo et al., 2016). (B) and (C) Gc is colored according to the convention for Class II fusion proteins: domains I, II, and III are shown in red, yellow, and blue, respectively. The DI–DIII linker is shown in cyan, and the beginning of the stem region at the C-terminus of the crystallized ectodomain constructs is shown in magenta. Disulfide bonds are shown as sticks, with sulfur atoms colored green. (C) The trimer is oriented axially to the viral and target membranes, with the fusion loops in domain II at the top of the structure.