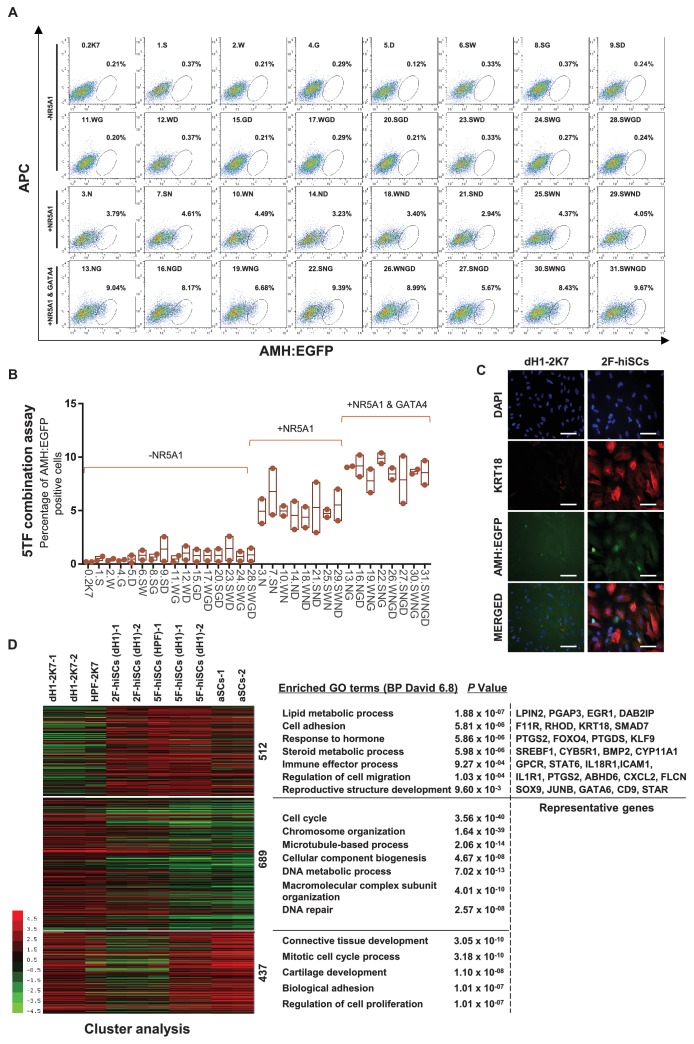
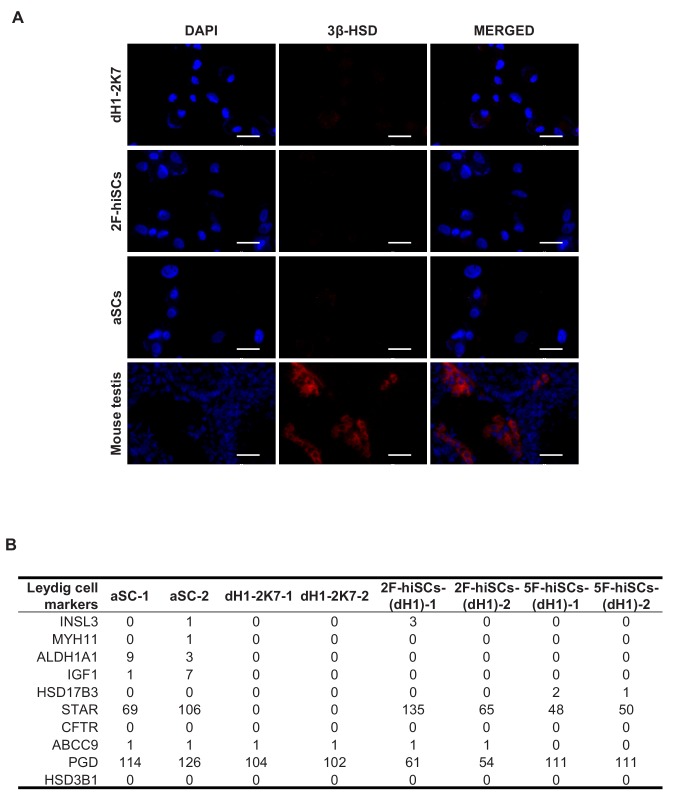
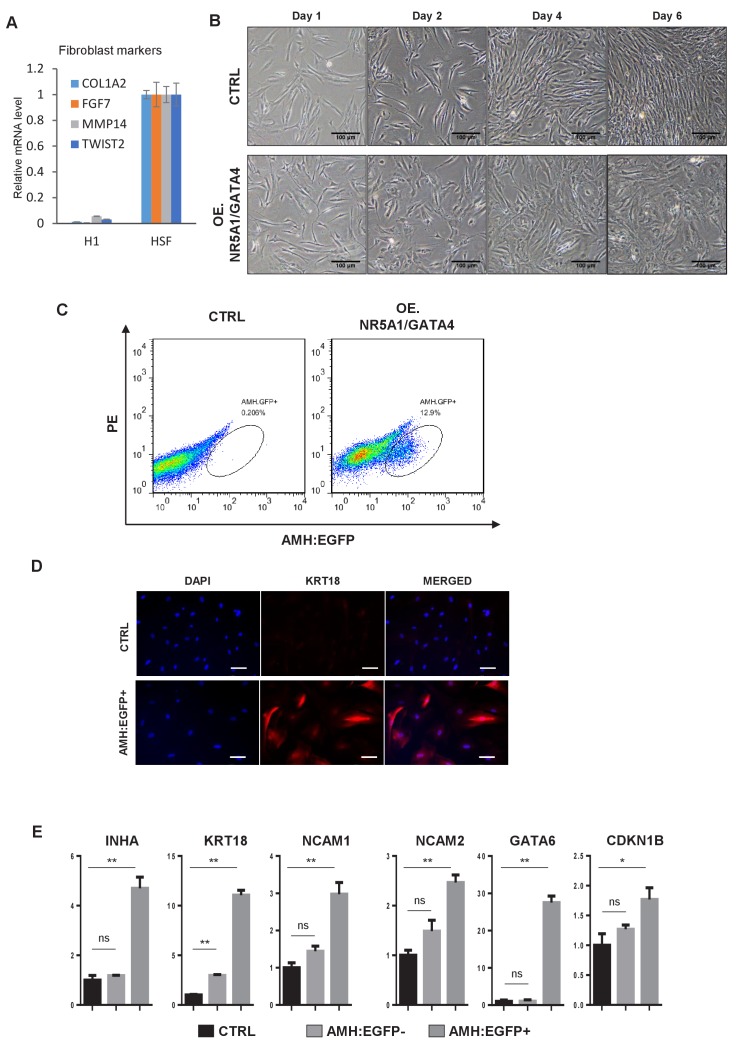
Figure 3. NR5A1 and GATA4 are sufficient to derive hiSCs.
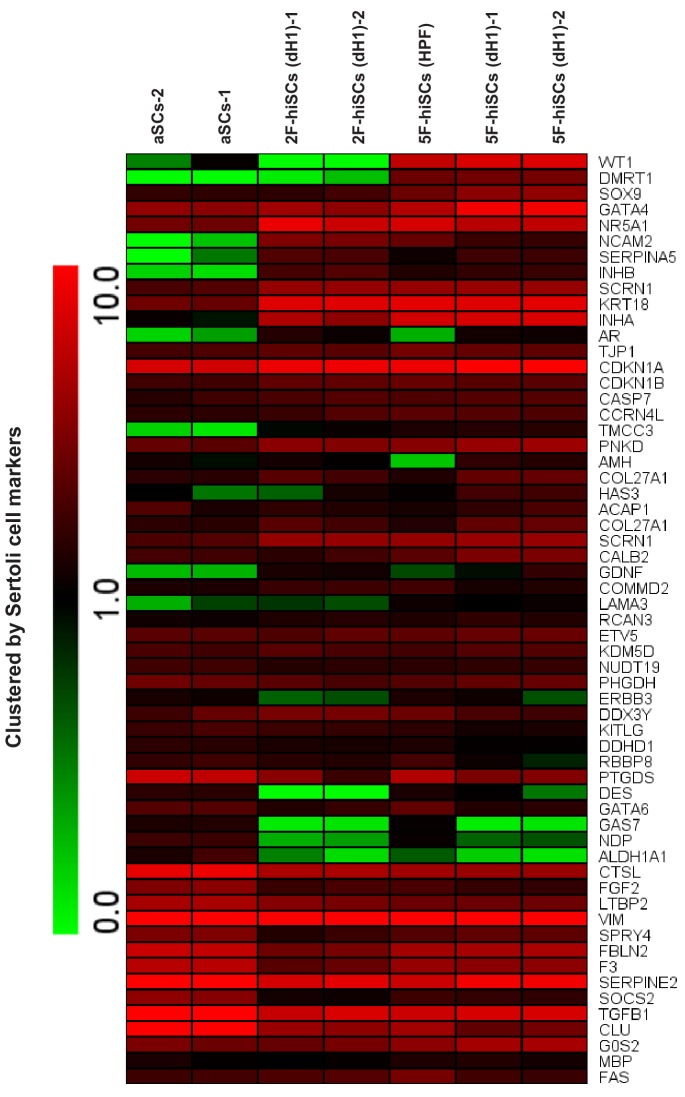
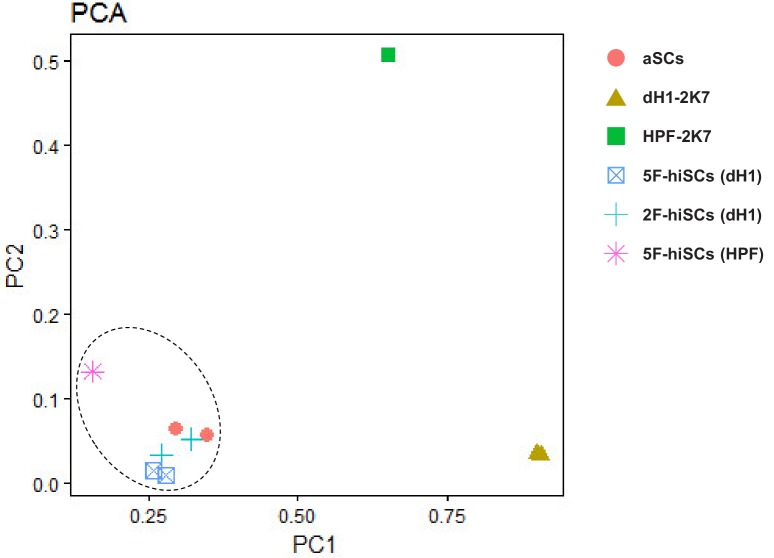
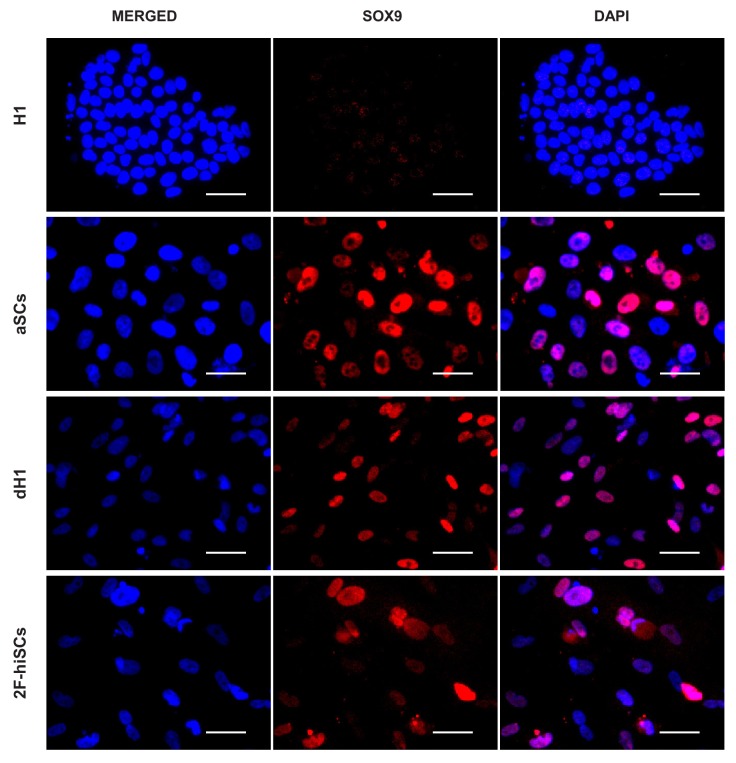
(A) Representative FACS results of different combinations of NR5A1, GATA4, WT1, SOX9 and DMRT1 for the induction of AMH:EGFP+ cells. dH1 fibroblasts were transduced with the indicated factors and reprogrammed for 10 days. The combinations were divided into three groups: -NR5A1, combinations without NR5A1; +NR5A1, combinations with NR5A1 but without GATA4; +NR5A1 and GATA4, combinations with both NR5A1 and GATA4. (B) Quantitative data of EGFP+ cells in (A). n = 2, biological replicates, error bar indicates SD, three independent combination experiments were conducted. ~104 cells were analyzed in each experiment. (C) Immunofluorescent staining of KRT18 in 2F-hiSCs and dH1-2K7 after FACS. Scale bar = 100 μm. (D) Heat map of RNA-seq data illustrating differentially co-expressed genes. Red indicates upregulated expression, whereas green indicates downregulated expression. The genes were divided into three groups: Genes upregulated in 2F-hiSCs (dH1), 5F-hiSCs (dH1) and aSCs; genes upregulated in 5F-hiSCs (dH1) and aSCs but downregulated in 2F-hiSCs (dH1); genes downregulated in 2F-hiSCs (dH1), 5F-hiSCs (dH1) and aSCs; all compared to dH1-2K7. The gene number of each group was listed next to the map. Functional enrichment terms of each group and the representative genes were shown on the right side.