Fig. 1. Nup53 uses distinct IDRs for binding to partner nups and Kap121.
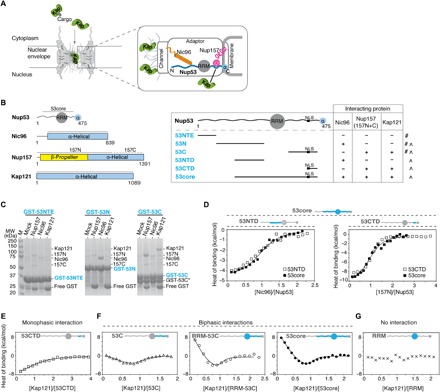
(A) A cross-sectional view of the NPC shows distinct nup subcomplexes linked by IDRs in adaptor nups (wavy lines). Kaps (green) facilitate NLS-mediated nuclear import. The zoomed-in section highlights the adaptor Nup157·Nup53·Nic96 complex that bridges the channel and membrane nup modules. (B) Domain architecture of Nup53, Nic96, Nup157, and Kap121 (left panel). Gray lines mark boundaries of the protein constructs used in this study. Table showing Nup53 fragments and the results of pull-down (#) and ITC (˄) binding assays (right panel). (C) Pull-downs using partially purified, GST-immobilized Nup53 fragments with Nic96, Nup157 (157N and 157C), or Kap121. Eluted GST·Nup53 complexes were visualized by SDS–polyacrylamide gel electrophoresis (PAGE) (M, molecular weight marker; GST-53C*, proteolytic truncation product). (D) ITC profiles for 53NTD-Nic96 and 53CTD-Nup157 interactions overlaid with Nic96 or 157N titrations with 53core. Integrated heat signals were analyzed by a single-site 1:1 binding model, yielding binding constants of 7.0 (± 2.2) × 105 M−1, 9.9 (± 0.9) × 105 M−1, 1.6 (± 0.2) × 106 M−1, and 1.1 (± 0.3) × 106 M−1 for 53NTD-Nic96, 53core-Nic96, 53CTD-Nup157, and 53core-Nup157 interactions, respectively. (E to G) ITC profiles for interactions of Kap121 (150 μM) with different Nup53 fragments (15 μM; blue). Monophasic binding data (E) were fitted with a single-site 1:1 binding model. The biphasic binding isotherms (F) were analyzed by either two-mode (53C) or multiple-equilibrium (RRM-53C and 53core) binding models (continuous lines). The best-fit parameters and the associated errors from global analysis are presented in Table 1. The RRM domain in Nup53 does not interact with Kap121 (G).
