Fig. 4. Karyopherins modulate molecular architecture of nucleoporin assemblies.
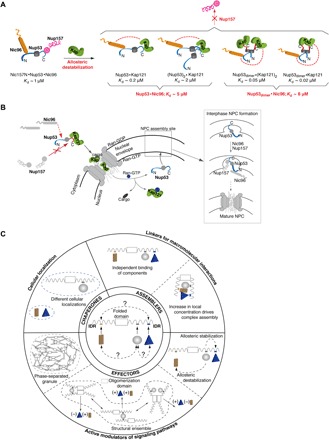
(A) Schematic representation of Nup53 hub interactions. The ternary complex containing Nup157, Nup53, and Nic96 is destabilized by Kap121 binding to the Nup53 C-terminal IDR. Kap121 allosterically reduces Nic96 affinity for the monomeric and dimeric conformations of the complex. Dissociation constants (Kds) for the different complexes were derived from Table 1. (B) Proposed model for allostery-driven Nup53 hub assembly in yeast. Kap121 binds to a cytoplasmic pool of Nup53 to destabilize its interactions with Nic96 and Nup157. In the nucleus, Ran-GTP releases Nup53. At the nuclear envelope, Nup53 sequentially recruits other adaptor nucleoporins. In the final step of NPC assembly, Nic96 recruits channel nucleoporins. (C) Classification of IDR-mediated interactions within macromolecular complexes. A hub protein consisting of a folded domain and IDR binding sites for three interacting partners is shown in the center. Different binding scenarios are considered in increasing order of complexity. By binding to chaperones (top left), IDRs target proteins to particular subcellular locations. As assemblers, IDRs independently recruit macromolecules or promote avidity-driven interactions. As effectors, IDRs are often regulated by cooperative binding and/or allostery, actively modulating biological activity of macromolecular complexes or signaling pathways. Coupling of folded and IDR domains may promote the formation of a structural ensemble or a phase-separated granule (dashed lines).
