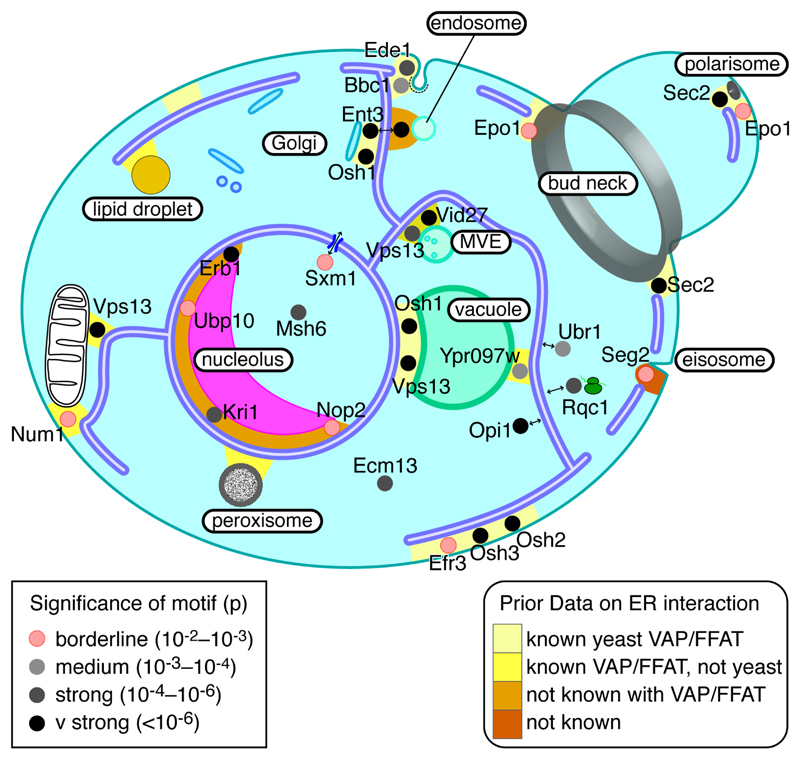
Figure 4. Cellular distribution of proteins with FFAT motifs.
Likely location of 24 yeast proteins with FFAT motifs, including 4 known, all 11 with FFAT scores ≤1.5, and a sample of 9 others with FFAT scores 2.0 or 2.5. Each protein is coloured by the significance of the Chi2 test of the number of motifs in its orthologue family over background (Table 2, and see Key): pink = borderline significance, increasingly dark grey = increasing significance. To reflect known localisation to multiple contact sites, Ent3, Epo1, Osh1, Sec2 and Vps13 appear more than once. Contacts between the ER and other cellular components are coloured according to the level of prior knowledge (see Key): light yellow (VAP-FFAT bridge known in yeast); dark yellow (VAP-FFAT bridge known but not in yeast); orange (contact known, but no VAP-FFAT bridge in any cell type); red (contact unknown). Abbreviation: MVE – multivesicular endosome. Note that no yeast FFAT proteins have been predicted yet that target lipid droplets or peroxisomes.