Abstract
KRAS is the most frequently mutated driver oncogene in human adenocarcinoma of the lung. There are presently no clinically proven strategies for treatment of KRAS-driven lung cancer. Activating mutations in KRAS are thought to confer independence from upstream signaling, however recent data suggest that this independence may not be absolute. Here we show that initiation and progression of KRAS-driven lung tumors requires input from ERBB family receptor tyrosine kinases (RTKs): Multiple ERBB RTKs are expressed and active from the earliest stages of KRAS-driven lung tumor development, and treatment with a multi-ERBB inhibitor suppresses formation of KRASG12D-driven lung tumors. We present evidence that ERBB activity amplifies signaling through the core RAS pathway, supporting proliferation of KRAS-mutant tumor cells in culture and progression to invasive disease in vivo. Brief pharmacological inhibition of the ERBB network enhances the therapeutic benefit of MEK inhibition in an autochthonous tumor setting. Our data suggest that lung cancer patients with KRAS-driven disease may benefit from inclusion of multi-ERBB inhibitors in rationally designed treatment strategies.
Introduction
Cancers of the lung account for over 1.5 million deaths per annum worldwide, and 5-year survival rates remain between 10 and 15% in many developed countries [1]. The majority of lung cancers are classified as non-small cell (NSCLC), and adenocarcinoma is the most common histological subtype of NSCLC. Activating mutations in KRAS occur in a third of lung adenocarcinoma (LuAd) cases [2]. RAS proteins have historically proven to be elusive targets for selective inhibition, although the recent development of G12 mutant KRAS-selective tool compounds suggests that therapeutic targeting of KRAS may in time be possible [3, 4]. In the interim, there is a pressing need to develop alternative strategies for more effective treatment of KRAS-driven disease.
The ERBB family of receptor tyrosine kinases is comprised of 4 members, EGFR (ERBB1), HER2 (ERBB2, NEU), ERBB3, and ERBB4, all of which can homo- or heterodimerize, and dimerization is required for signaling activity. ERBB dimers are activated upon binding a spectrum of soluble ligands including EGF, epiregulin (EREG), amphiregulin (AREG), and neuregulin (NRG), amongst others, together forming a network for ERBB-driven signal transduction [5]. EGFR is a well-recognized driver of lung adenocarcinoma, with genetic alterations present in up to 18% of cases [2]. ERBB2 and ERBB3 are highly expressed in embryonic lungs of humans and rodents, and expression of both persists into adulthood [6, 7]. Overexpression of ERBB2 in the absence of gene amplification is common in human LuAd [8, 9], and functionality of ERBB2/ERBB3 heterodimers in NSCLC-derived cell lines was previously shown [10]. Amplification of any of the 4 ERBB RTKs is associated with poor prognosis in lung cancer [11], and high expression of the promiscuous ERBB ligand EREG has previously been linked to disease progression and aggressive phenotypes in models of EGFR- and KRAS-driven lung cancer [12, 13].
In a wild-type setting, ligand-activated signaling through ERBB RTKs activates KRAS [14]. Mutation of KRAS is generally thought to confer independence from upstream regulation, a view that is reinforced by the mutual exclusivity of activating mutations in KRAS and EGFR in LuAd, and by the failure of EGFR-selective inhibitors to show therapeutic benefit against KRAS-driven cancers [15, 16]. However, several recent results suggest that the independence of mutant KRAS from upstream signaling may not be absolute: In KRAS-mutant NSCLC cell lines, activation of PI3K is contingent upon basal activity of wild-type IGFR, establishing an important precedent for coordination of oncogenic and normal signal transduction [17]; genetic deletion of EGFR was shown to suppress development of KRASG12D-driven pancreatic ductal adenocarcinoma [18, 19]; induced expression of ERBB2 and ERBB3 was found to underlie resistance of KRAS-mutant lung and colorectal cell lines to MEK inhibition [20]. Strikingly, in the latter study, MEK inhibitor-induced ERBB2/3 expression was associated with recovery of ERK phosphorylation downstream of KRAS, suggesting a surprising role for upstream signaling in sustaining pathway activity despite the presence of activated KRAS.
We therefore examined the requirement for ERBB activity in an inducible model of progressive autochthonous LuAd, driven by the combination of endogenously expressed KRASG12D and modest overexpression of c-MYC. We present evidence that redundant signal transduction through multiple ERBB RTKs supports development and progression of mutant KRAS-driven lung tumors. Our data suggest that front-line use of multi-ERBB inhibitors may show clinical benefit in KRAS-driven LuAd.
Results
ERBB activity is required for KRasG12D-driven lung tumor formation
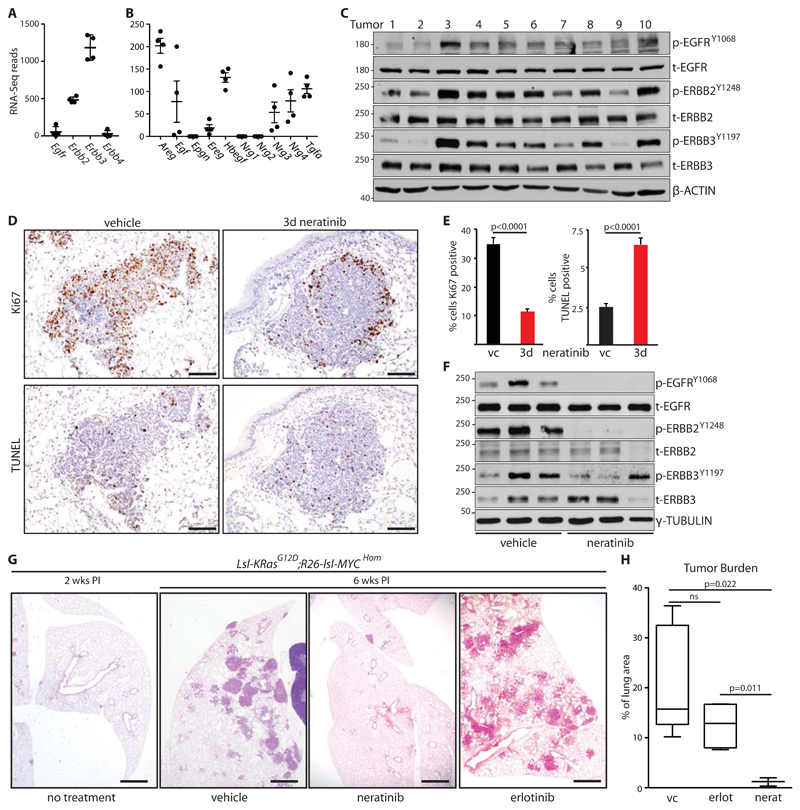
Induced expression of ERBB-family receptor tyrosine kinases (RTKs) is associated with resistance of KRAS-mutant NSCLC cell lines to MEK inhibition [20]. We therefore examined expression of ERBB RTKs and their ligands in micro-dissected early-stage lung tumors, using a CRE-inducible mouse model of autochthonous lung adenocarcinoma driven by KRASG12D combined with modestly increased MYC (lsl-KRasG12D;Rosa26-lsl-MYC – henceforth KM), the latter expressed from the Rosa26 locus in amounts that alone fail to provoke a phenotype (fig. S1A). In tumor samples harvested 6 weeks after allele activation, we found strong expression of Erbb2 and Erbb3 mRNA, whereas Egfr was weakly expressed, and Erbb4 was not detected in tumors from 2 of 4 KM mice (Fig. 1A). Multiple ERBB ligands were expressed, with Areg, Tgfα and Hbegf showing strongest expression, while Egf, Ereg, Nrg3, and Nrg4 were also clearly detected (Fig. 1B). The presence of both RTKs and multiple cognate ligands suggested that ERBB RTKs may actively signal in developing KRASG12D-driven lung tumors. Lysates prepared directly from multiple individual tumors, harvested 6 weeks after induction and immunoblotted for phospho-ERBB RTKs, consistently showed readily detectable phospho-EGFR, phospho-ERBB2, and phospho-ERBB3, suggesting that these RTKs are indeed active in developing KRASG12D-driven lung tumors (Fig. 1C & fig. S1B). To determine if ERBB signaling functionally contributes to tumor development, we treated tumor-bearing mice acutely with a multi-ERBB inhibitor, neratinib [21]. Treatment of mice for 3 days with neratinib suppressed activity of ERBB RTKs, reduced tumor cell proliferation, and increased apoptosis, suggesting that ERBB activity promotes growth of KRASG12D-driven tumors (Fig. 1D-F). Continuous daily treatment of mice from 2 weeks after allele induction almost completely suppressed the emergence of tumors, indicating that the combination of endogenously expressed KRASG12D and constitutively expressed MYC requires input from ERBB RTKs in order to give rise to lung tumors (Fig. 1G, H). In contrast, daily erlotinib treatment failed to show the same effect, consistent with reports that EGFR inhibition in isolation is ineffective in KRAS-driven lung cancer [15, 18].
Figure 1. ERBB activity is required for KRAS-driven lung tumor formation.
A) Expression of ERBB family RTKs in KM lung tumors harvested 6 weeks post allele induction (PI), measured by RNA-SEQ. Mean±SD read counts in tumors from 4 mice shown. B) Expression of ERBB family ligands in KM lung tumors harvested 6 weeks PI, as per (A). C) Immunoblots of lysates generated from 10 individual KM tumors, harvested 6 weeks PI, using the indicated antibodies. D) Representative images of IHC for Ki67 and TUNEL staining of KM mice treated for 3 days with 80 mg/kg neratinib (n=3) or vehicle control (n=3). Scale bars = 100 μm. E) Quantification (mean ± SEM) of staining in 5 tumors from each mouse as per (D). Left panel shows % of tumor cells expressing Ki67; right panel shows % of TUNEL-positive tumor cells; vc = vehicle control. P values are from 2-tailed T tests. F) Immunoblots of 3 individual KM tumors from mice treated for 3 days with neratinib or vehicle control. G) Representative H&E images from KM mice treated daily with neratinib (2 x 40 mg/kg) or erlotinib (2 x 50 mg/kg), commencing 2 weeks PI, and harvested at 6 weeks PI. Scale bar = 1 mm. H) Quantification of tumor burden from (G). Box and whisker plots show median, interquartile and 99% range of tumor area, expressed as a percentage of total lung tissue area, measured across >25 sections from each mouse. N = 5 vehicle control (vc); n = 4 erlotininb; n = 3 neratinib. ANOVA followed by Tukey test, ns=not significant.
Progression of KRas-driven lung tumors is associated with increased ERBB network expression
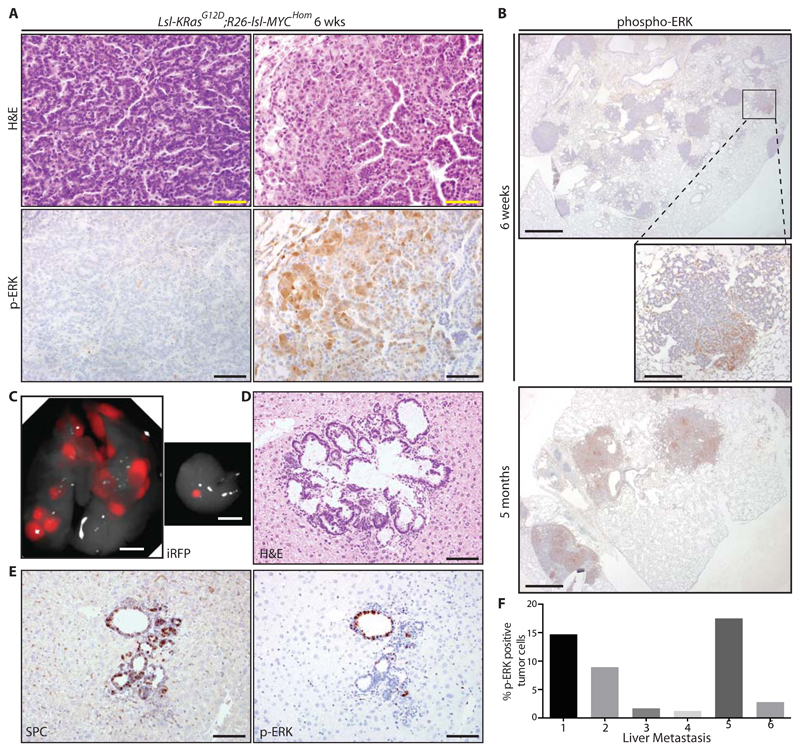
The above analysis was performed on tumors at 5-6 weeks after adeno-CRE-mediated allele activation. At this time after induction, the lungs of all induced KM mice contained dozens of individual tumors that presented with uniform histology resembling human papillary adenocarcinoma in situ (Fig. 2A, left panels). A small minority of tumors (2-5%) also contained a second, more disorganized population with more aggressive histological features, including increased cytosolic and nuclear volume, prominent nucleoli, and increased morphological heterogeneity (Fig. 2A, right panels). IHC analysis revealed continuous expression of the type II pneumocyte marker SPC across both populations in individual tumors, suggesting that the second population represents the emergence of aggressive sub-clones (fig. S1C). Progression of KRASG12D-driven lung tumors to more aggressive disease is associated with a pronounced increase in ERK1/2 phosphorylation [22, 23]. Accordingly, IHC analysis of phospho-ERK (p-ERK) revealed much higher expression of p-ERK in these disorganized sub-clones compared with the rest of the same tumors, and indeed across the entire tumor population (Fig. 2A & B). By comparison, KM tumors harvested at 4-6 months after induction showed widespread expression of p-ERK, because these more aggressive sub-clones gradually came to predominate (Fig. 2B, bottom panel). Moreover, KM metastases to the liver also show high amounts of epithelial p-ERK staining (Fig. 2C-F).
Figure 2. KM lung tumor progression is associated with increased ERK phosphorylation.
A) Images of H&E (upper panels) and phospho-ERK (lower panels) stained KM lung tumors harvested at 6 weeks PI illustrating histological changes associated with tumor progression: left panels are representative of >95% of total tumor area at 6 weeks PI, while right panels represent 2-5% of total tumor area at 6 weeks PI. Scale bar = 50 μm. B) Phospho-ERK staining in KM tumors harvested at 6 weeks PI (top and center panels) versus 5 months PI (bottom panel). Scale bars = 1 mm (top and bottom panels) and 200 μm (center panel). C) Detection of Hprt-lsl-IRFP expression in primary lung tumors (left) and a liver metastasis (right) in a KM mouse harvested 6 months PI. Scale bar = 5mm. D) Histological confirmation of liver metastasis stained by H&E. E) IHC detection of SPC and p-ERK in the same metastasis as (D). Scale bar = 50 μm (D and E). Images are representative of 6 mice. F) Halo quantification of p-ERK positive cells in individual metastases, expressed as % of tumor cells.
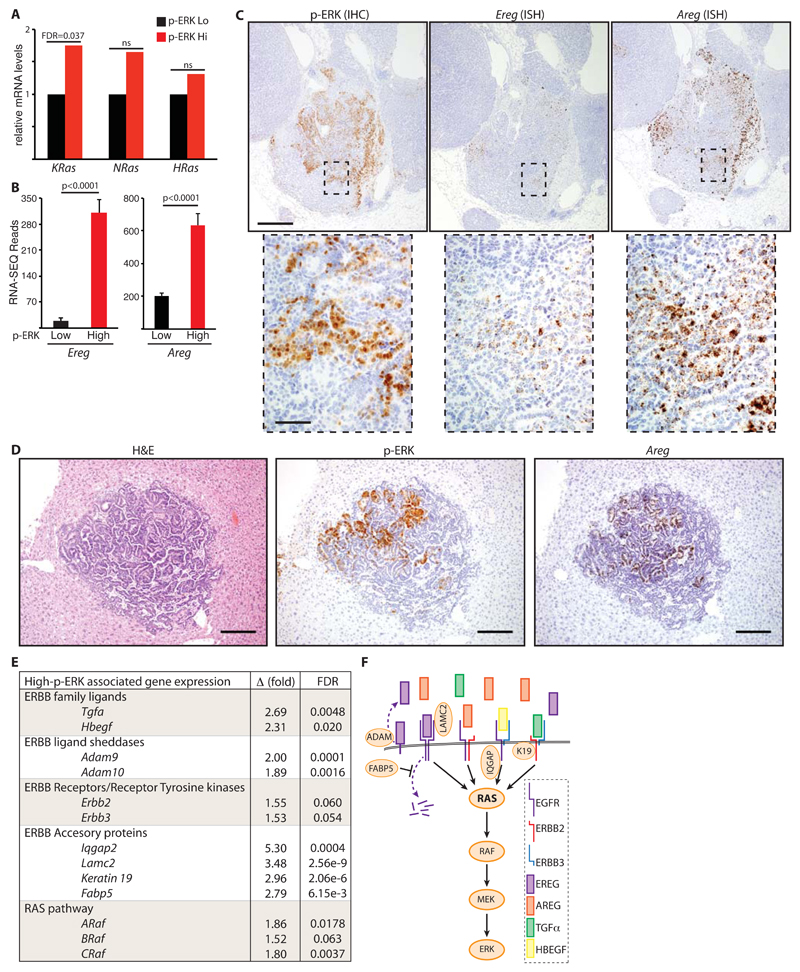
Increased p-ERK was previously shown to be associated with amplification of the mutant KRas locus in a KP lung cancer model wherein KRASG12D expression was combined with loss of functional p53 [24]. We used laser-capture micro-dissection coupled with RNA-SEQ analysis to compare gene expression in p-ERKLow with p-ERKHigh KM tumor regions from 4 individual mice (see schematic, fig. S1D). Expression of KRas was modestly higher (< 2 fold) in pERKHigh tumor regions, suggesting that locus amplification is not the underlying driver of progression in the KM model (Fig. 3A). However, we detected a sharp increase in expression of promiscuous ERBB-family ligands, Ereg and Areg, along with more modest increases in Hbegf and Tgfα (Fig. 3B-D and fig. S1E). Increased expression of Ereg and Areg was previously reported to be associated with more aggressive tumors in an inducible model of KRAS-driven mammary cancer [25] and in human NSCLC cells enriched for metastatic behavior [26]. In situ hybridization revealed a clear overlap between expression of Ereg, Areg and extensive phosphorylation of ERK (by IHC) in tumor epithelium, confirming the RNA-SEQ data (Fig. 3C and fig. S2A & B). The same phenotype of focal progression to p-ERKhigh accompanied by dramatically increased Areg expression was also observed in primary lung tumors upon induction with a more lineage-restricted adeno-SPC-CRE (fig. S2A & B), and in liver metastases after induction with the standard adeno-CMV-CRE (Fig. 3D and fig. S2C), arguing against the possibility that this phenotype might arise from inadvertent activation of the driver oncogenes in other lung-resident populations [27]. Additionally, RNA-SEQ analysis revealed increased expression of Adam9 and Adam10 sheddases, which process membrane-bound ERBB ligands into their more potent soluble forms [28]; increased expression of the signaling scaffold Iqgap2; and increased expression of LamC2, Fabp5, and Keratin19, all recently shown to enhance signaling through EGFR/ERBB family RTKs [26, 29, 30]. Expression of Erbb2 and Erbb3 was not significantly changed in p-ERKHigh regions (Fig. 3E & F).
Figure 3. Expression of the ERBB network increases during progression from p-ERKlow to p-ERKhigh KM tumors.
A) Normalized expression of RAS genes in laser-capture micro-dissected (LCM) p-ERKHigh KM tumor regions relative to p-ERKLow regions from tumors in the same mice (n = 4 mice), measured by RNA-SEQ. False discovery rate (FDR) shown for KRAS; ns = not statistically significant. B) Mean and SEM RNA-SEQ reads of Ereg and Areg mRNA from p-ERKLow & p-ERKHigh KM tumor regions from 4 mice. Adjusted p values were calculated in R. C) Serial sections of KM tumors stained by IHC for p-ERK (left panels) or by in situ hybridization for Ereg (center panels) or Areg (right panels). Scale bars = 200 μm (upper panels) & 25 μm (lower panels). D) Representative images of p-ERK (IHC) and Areg (ISH) expression in KM liver metastasis. Scale bar = 200 μm. Images are representative of 4 metastases with sufficient material. E) Normalized expression of ERBB network genes showing mean fold increase (Δ) in expression in p-ERKHigh relative to p-ERKLow KM tumor regions from 4 mice as per (A). FDR = false discovery rate. F) Diagrammatic representation of up-regulated components of the ERBB-RAS-ERK pathway associated with KM tumor progression.
These data suggest an alternative route to increased RAS pathway signaling that does not require KRas amplification but rather involves increased activity of the ERBB network, defined here as consisting of ERBB ligands, RTKs, and RTK accessory molecules such as LamC2 and Fabp5. Notably, examination of the TCGA lung adenocarcinoma (LuAd) dataset via cBioPortal [31] revealed overexpression and/or amplification of one or more constituents of the ERBB network in the majority of KRAS-mutant human LuAd (fig. S3).
ERBB signaling amplifies RAS pathway activity in KRAS-mutant human NSCLC cells
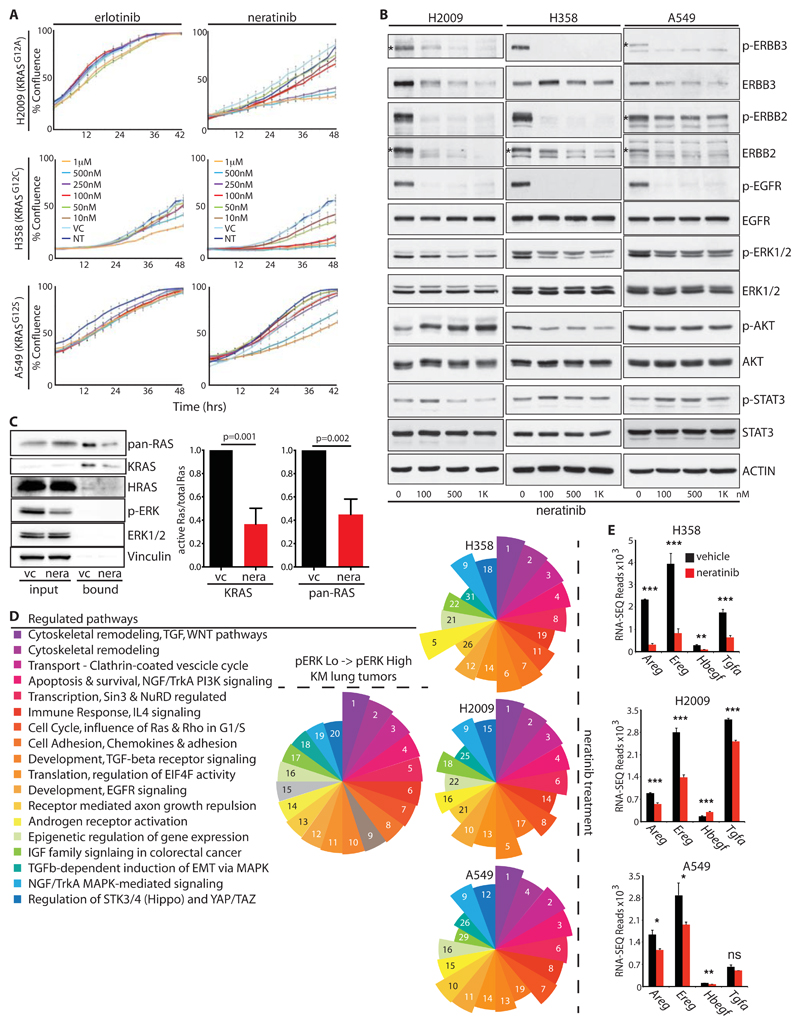
Combined inhibition of both EGFR and ERBB2 is required to prevent outgrowth of MEK inhibitor-resistant clones of human KRAS-mutant NSCLC cells, however the effect of multi-ERBB inhibition on treatment-naïve cells was not previously explored [20]. Treatment of multiple KRAS-mutant human NSCLC lines with the multi-ERBB inhibitor neratinib suppressed proliferation in a dose-dependent manner (Fig. 4A). In contrast, inhibition of EGFR in isolation showed little effect. Immunoblotting confirmed that the doses of neratinib used strongly suppressed activity of ERBB RTKs (Fig. 4B). Consistent with previous reports that neratinib drives increased ERBB turnover [32], ERBB2 and ERBB3 protein expression was also reduced by neratinib in multiple cell lines. Despite the presence of G12 mutant KRAS in all such cells neratinib consistently reduced phospho-ERK whereas effects on AKT and STAT3 varied across the cell lines tested (Fig. 4B & fig. S4A). We therefore examined RAF binding as a direct measure of RAS signaling activity in A549 cells, which are homozygous for mutant KRAS. Acute ERBB inhibition reduced both total RAS:RAF & specific KRAS:RAF binding by >50%, consistent with the observed partial reduction in p-ERK (Fig. 4C). Additionally, murine lung tumor cells carrying a spontaneously amplified KRasG12D allele also showed sensitivity to neratinib, albeit somewhat lower than that of cells carrying a single copy of the allele (fig. S4B), and RAS-less MEFs reconstituted with G12D mutant KRAS were likewise sensitive to neratinib, albeit less so than those reconstituted with wt KRAS (fig. S4C & D). Together these results suggest that ERBB amplifies signaling through the core RAS->ERK module, even in cells with activating mutation of KRAS, whereas sensitivity to ERBB suppression is imparted through both mutant and wild-type KRAS.
Figure 4. A feed-forward ERBB signaling loop drives proliferation of KRAS-mutant human NSCLC cells.
A) Growth curves of 3 KRAS-mutant human NSCLC lines upon treatment with increasing doses of the EGFR-selective inhibitor erlotinib or the multi-ERBB inhibitor, neratinib, measured by Incucyte time-lapse video-microscopy. Error bars show SD for technical triplicates. Data are representative of at least 2 independent experiments. B) Lysates from KRAS-mutant NSCLC cells treated with increasing doses of neratinib, immunoblotted with the indicated antibodies. Asterisks, where present, indicate the correct band. C) RAS immunoblots of RAF-coated bead precipitates from lysates of A549 cells treated with neratinib or vehicle control for 2 hours. Lysate input aliquots were also immunoblotted with the indicated antibodies. Right panel shows mean ± SEM for quantification of KRAS band intensities from 3 independent experiments (arbitrary units). P values are from 2-tailed T-tests. D) Top 20 significantly modulated pathways associated with the transition to p-ERKHigh disease in the KM model, identified using Metacore GeneGO analysis of RNA-SEQ expression data. Segment size in the pie chart (left panel) reflects ranking of the pathways by false discovery rate (FDR). Right panels show that 18 of the same pathways are modulated in each of 3 KRAS-mutant human NSCLC lines after overnight treatment with neratinib (250nM for A549 and H2009; 25nM for H358). Numbers and pie segment size reflect ranking by FDR. E) Expression of ERBB ligands in the indicated cells treated overnight with vehicle (black) or neratinib (red), measured by RNA-SEQ as per (D). Mean & SEM of biological triplicates shown. P values are from 2-tailed T-tests, abbreviated as follows: * =<0.05; ** = <0.01; *** <0.0001; ns = not significant.
To gain further mechanistic insight into the growth inhibitory effects of neratinib, we performed unbiased transcriptomic analysis after overnight treatment of 3 KRAS-mutant NSCLC cell lines. Metacore GeneGO pathway analysis revealed a remarkable degree of consistency across the 3 cell lines tested. Moreover, 18 of the 20 most highly modulated pathways associated with progression of murine KM tumors to p-ERKHigh disease were reciprocally modulated in the human cells upon neratinib treatment (Fig. 4D & Table S1). In all three cell lines, ERBB inhibition reduced expression of the same ERBB ligands that increased as KM tumors progressed to p-ERKHigh disease (Fig. 4E). These findings indicate that ERBB activity establishes a feed-forward loop that sustains KRAS-mutant NSCLC proliferation in vitro and drives tumor progression in vivo, at least in part by amplifying signaling through the core RAS-ERK module.
ERBB inhibition enhances the potency of MEK inhibition in vitro and extends survival of mice with LuAd
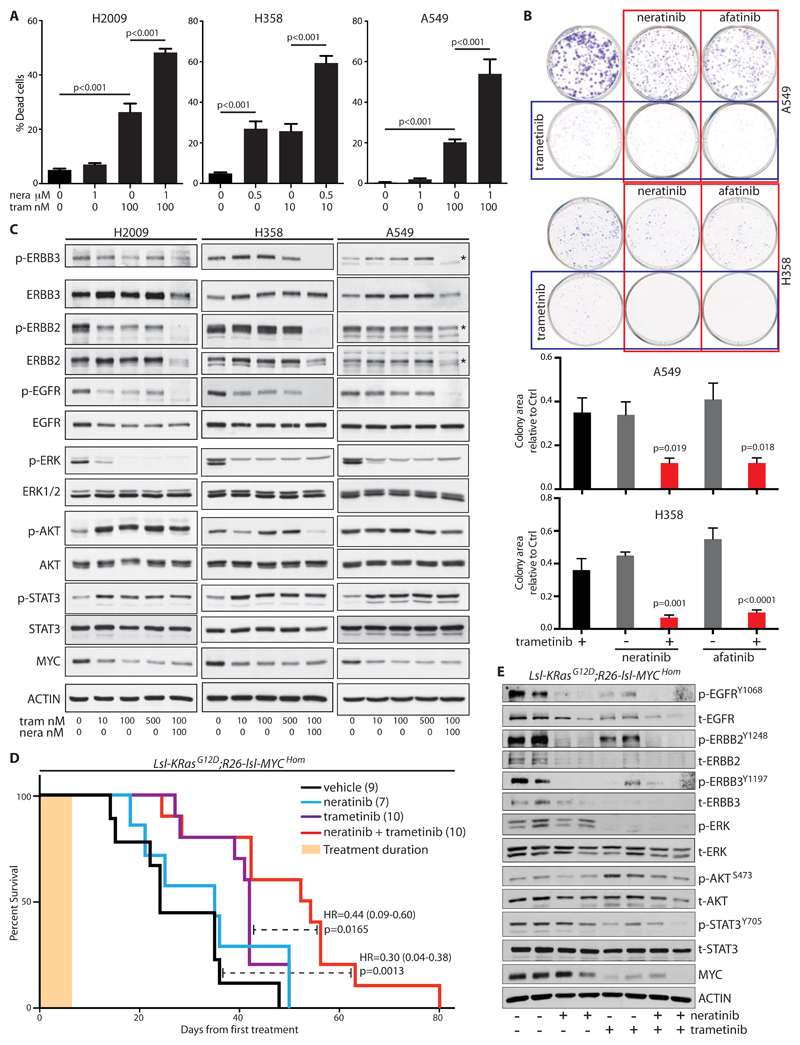
With the exception of H358 cells, ERBB inhibition alone did not result in death of KRAS mutant NSCLC cells in vitro, however, neratinib increased apoptosis induced by inhibition of MEK downstream of KRAS in multiple cell lines (Fig. 5A). Substitution of neratinib with a second multi-ERBB inhibitor, afatinib, replicated the effects of neratinib, confirming the on-target specificity of the drug (fig. S5A). Moreover, both drugs combined with MEK inhibition to suppress colony formation (Fig. 5B). Consistent with published results [20], MEK inhibition alone increased expression of ERBB2 and ERBB3 in vitro, although the effect on ERBB2 phosphorylation was variable across the cell lines tested (Fig. 5C). Co-treatment with neratinib suppressed both expression and activity of ERBB2 and ERBB3, while also suppressing EGFR activity (Fig. 5C). Downstream of the RTKs, trametinib alone consistently suppressed expression of p-ERK and c-MYC, but increased phosphorylation of AKT and STAT3 (Fig. 5C). Suppression of increased AKT phosphorylation by neratinib was clear only in H358 cells, but the drug combination failed to reduce STAT3 phosphorylation. The growth inhibitory effects of the drug combination in vitro thus better correlate with suppression of the core RAS-ERK-MYC pathway than with the activity of ancillary signaling pathways.
Figure 5. ERBB blockade enhances MEK inhibitor-driven apoptosis in vitro and therapeutic impact in vivo.
A) Apoptosis induced in human NSCLC cells, measured 48 hours after treatment with the indicated doses of neratinib (nera) and/or trametinib (tram). Mean ± SEM of 3 independent experiments shown (ANOVA & Tukey test). B) Clonogenic assay showing suppression of colony formation in A549 and H358 cells after 48 hours of treatment with the indicated inhibitors. Lower panels show quantification of colony area (% surface coverage) from 5 independent experiments. Significance was determined for drug combinations versus trametinib alone. (ANOVA & Tukey test). C) Lysates from the indicated cells treated for 24 hours with increasing doses of trametinib alone or the combination of trametinib and neratinib, immunoblotted with the indicated antibodies. Asterisks, where present, indicate the correct band. D) Overall survival, measured from the first day of treatment, of tumor-bearing KM mice treated daily for 1 week (tan bar) with neratinib (80 mg/kg), trametinib (1 mg/kg), or the combination of both, then followed without further intervention. Treatment was commenced at 5 weeks PI. Cohorts shown are vehicle (n=9); neratinib (n=7); trametinib (n=10); trametinib + neratinib (n=10). Logrank hazard ratios (HR±95% CI) and p values are shown for comparisons of T+N versus vehicle and T+N versus T alone (dashed lines). E) Lysates of individual tumors from mice treated with neratinib (80 mg/kg) and/or trametininb (1 mg/kg) for 3 days, blotted with the indicated antibodies.
The combination of MEK and multi-ERBB inhibition was previously shown to suppress the growth of subcutaneous NSCLC xenografts under continuous daily treatment [20], however, cell line xenografts have a poor track record of accurately predicting human patient responses. We therefore tested the therapeutic efficacy of transient inhibition of ERBB and/or MEK in our fully immunocompetent KM model of autochthonous lung adenocarcinoma. Tumors were induced in adult KM mice and allowed to develop for 6 weeks. The presence of an IRFP Cre-reporter allele [33] in a subset of such animals allowed us to confirm the presence of tumors by Licor PEARL fluorescence imaging before initiating therapy (fig. S5B). Mice were treated daily for 1 week with neratinib, alone or in combination with trametinib, then left untreated to determine the effect on overall survival. Transient ERBB blockade alone showed little influence on overall survival, whereas MEK inhibition alone extended survival of KM mice. The combination of neratinib and trametinib further extended survival over that achieved by MEK inhibition alone (Fig. 5D). Fluorescence imaging of IRFP-positive KM mice showed pronounced suppression of lung tumor growth in individual mice treated with combination therapy (fig. S5B & C). Immunoblotting of tumors from mice treated for 3 days with either drug alone or both combined confirmed the pathway-specific effects of these inhibitors in vivo (Fig. 5E). We conclude from these data that multi-ERBB inhibition may benefit LuAd patients with mutant KRAS-driven disease, if used in combination with other agents such as MEK inhibitors.
Discussion
EGFR-selective inhibitors have failed to show clinical benefit in mutant KRAS-driven cancers. In contrast with targeted inhibition of EGFR in isolation, we show here that broad inhibition of the ERBB network, using the multi-ERBB inhibitor neratinib, almost completely suppresses formation of KRASG12D-driven lung tumors, and enhances the benefit of MEK inhibition in established tumors. We show that ERBB activity enhances signaling through the core RAS->ERK pathway, establishing a feed-forward loop that is associated with progression to invasion and metastasis. We also show that brief, transient inhibition of ERBB signaling enhances the therapeutic benefit of MEK inhibition in the autochthonous setting. Although this latter result is broadly supportive of the clinical implications of an earlier study showing that increased ERBB activity underlies resistance to MEK inhibition [20], our data are fundamentally distinct in that we demonstrate a clear role for ERBB activity from the very outset of KRAS-driven tumor initiation, as opposed to in reaction to targeted inhibition of MEK.
Mechanistically, we show that nascent KRasG12D-driven lung tumors express multiple ERBB ligands and that production of these ligands increases dramatically as tumors progress to more aggressive disease. In the cell culture setting, the release of these ligands drives autocrine signaling that supports the viability of cancer cells. In vivo, the release of such ligands into the extracellular milieu likely drives both autocrine and paracrine signal transduction in any local cell population that expresses ERBB RTKs. As such, it is plausible to consider that some of the tumor-suppressive benefit of neratinib may derive from inhibition of ERBB signaling in such stromal populations. Nevertheless, our cell culture analysis argues that the primary effect of ERBB inhibition is indeed autonomous to the tumor epithelium. Further work will be needed to determine if paracrine activation of ERBB RTKs on attendant stromal cell populations plays any role in tumor maintenance or progression.
Previous work has revealed that threshold levels of KRAS signaling are required for tumor initiation and progression: In an inducible model of HRASG12V overexpression in mammary glands, focal tumors arising spontaneously from non-transformed epithelium that expressed low amounts of the transgene exhibited a pronounced increase in expression of both the HRasG12V transgene and of multiple ERBB ligands, including Ereg, Tgfa, and Hbegf, again suggestive of feed-forward signal amplification [25]. More recently, this same “activity threshold” principle was shown to apply to ERK signaling as the key oncogenic effector pathway of KRAS in the lung [34]. Through pharmacological enhancement of ERK activity, this study showed that different airway cell types require distinct amounts of ERK signaling for oncogenic transformation by mutant KRAS, and that a second, higher, threshold signal was needed for progression to carcinoma. The existence of this higher threshold in KRAS-driven LuAd was already clear from the pronounced increase in ERK phosphorylation associated with tumor progression [25] which, in the KP model, is driven by spontaneous amplification of the G12D mutated allele [24]. Our data suggest that ratcheting up ERBB signal transduction provides an alterative route to RAS pathway signal amplification, independent of KRAS gene amplification. It is important to note that, from the human data, there does not appear to be a preferred mechanism of ERBB signal enhancement – overexpression of ligands, amplification of RTKs, and/or accessory molecules could all plausibly achieve the same effect. What is clear from these data is that the potential to increase ERBB signaling appears to be widespread in KRAS-mutant human LuAd.
Perhaps the most surprising observation is that endogenously expressed KRASG12D requires basal signaling from ERBB RTKs to initiate lung tumors, even when accompanied by MYC overexpression. We previously showed that expression from the Rosa26 locus is refractory to growth factor signaling [35] – thus the requirement for ERBB activity does not reflect an artificial need for ERBB to sustain expression of the MYC transgene, whereas the observed effects on total MYC expression likely reflect the influence of RAS signaling on MYC protein stability [36]. Instead, these data suggest that G12 mutant KRAS requires a push from upstream RTKs to breach the initial threshold needed for tumor initiation. Supporting this hypothesis, it was recently shown that G12 mutant KRAS, although no longer responsive to RAS-GAPs, does retain some level of intrinsic GTPase activity and thus cycles slowly between on and off states [4], opening the possibility for upstream signaling to influence activity, either directly or by promoting activity of RAS guanonucleotide exchange factors (GEFs). Alternatively, tonic signal transduction through wild-type RAS isoforms may need to combine with that from mutant KRAS to likewise breach the threshold for transformation [37]. Our data presently do not distinguish between these possibilities and, indeed, they may not be mutually exclusive.
In the course of preparing this manuscript we became aware of an independent study that strongly complements our observations and underscores our conclusions: The work by Moll and colleagues similarly demonstrates a requirement for ERBB signaling to support progression of KRASG12D-driven lung cancer in vivo [38]. Their study used an independent pan-ERBB inhibitor, afatinib, in the context of both KRASG12D-only and KRASG12D;p53Fl/Fl -driven tumor models. The striking similarities in the two studies attest to the on-target specificity of the 2 drugs used, while also showing that the effects are independent of the genetic strategy used to accelerate KRAS-driven disease and are therefore likely to have broader potential application.
Activating mutations in KRAS are currently considered grounds for exclusion from clinical treatment with EGFR/ERBB inhibitors, and single-agent trials of the same drugs have only demonstrated efficacy in patient cohorts with activating mutations in EGFR or ERBB amplification [39]. Indeed, our data concur that multi-ERBB inhibitors are unlikely to benefit KRAS-driven cancer patients if used in isolation. However, we show here the potential for such drugs to sensitize autochthonous KRAS-driven tumors to additional therapeutic agents, in this case to the MEK inhibitor trametinib. Our data argue that clinical use of multi-ERBB inhibitors as part of an inhibitor cocktail to treat KRAS-driven LuAd deserves re-examination.
Materials and Methods
Study Design
Rationale for the project was to investigate the mechanistic basis of tumor progression in early stage KRAS-driven lung cancer. Unbiased RNA-SEQ analysis identified increased expression of the ERBB network, prompting further mechanistic investigation. All mice were asymptomatic at the date of treatment initiation and were allocated to treatment groups randomly. For survival benefit analysis, the absence of prior data precluded a precise power calculation: Sample sizes were therefore estimated according to Lehr’s quick formula at 80% power {n=16(c.v.)2(ln[r.m.])2}, assuming a ratio of the means of 0.2 and a coefficient of variation of 0.3. For all other analyses, sample sizes were determined in accordance with the 3Rs. Animals were treated by facility staff without knowledge of anticipated outcomes. Humane endpoints were predefined as impaired breathing and/or hunching with 10% or more weight loss. Histology, IHC and ISH were performed blindly by the Beatson Institute histology core facility and were scored without reference to genotype or treatment group. Reporting is consistent with the ARRIVE guidelines. In vitro experiments were all performed on 3 separate occasions, except where noted. In vitro analyses were not blinded.
Genetically Engineered Mice & Mouse Procedures
Procedures involving mice were performed in accordance with Home Office license numbers 60/4183 & 70/7950 (CRUK BICR, UK). LSL-KRasG12D (B6.129S4-Krastm4Tyj/J) mice [40] were obtained from the NCI mouse repository at Frederick, MD, USA. Rosa26DM.lsl-MYC mice were generated as previously described [35] with the ATG-initiated human c-MYC (MYC2) cDNA, devoid of 5' and 3' UTRs, replacing the MYC-estrogen receptor ligand-binding domain fusion cDNA. Targeted insertion into the Rosa26 locus was confirmed by Southern blotting, and genotyping was initially performed with the following primers: A) CCC AAA GTC GCT CTG AGT TG (common); B) GCG AAG AGT TTG TCC TCA ACC (targeted locus); C) GGA GCG GGA GAA ATG GAT ATG A (wild-type locus). All genotyping was subsequently performed by Transnetyx Inc. Detailed in vivo procedures are provided in the supplemental material along with immunohistochemical, laser-capture microdissection and RNA-SEQ methodology.
Cell Culture and Related Assays
Human lung cancer cell lines (A549, H2009, H358) were validated in-house and grown in RPMI with 10% FBS and penicillin (100 units/ml)/streptomycin (100μg/ml). RAS-less MEFs reconstituted with wild-type or mutant KRAS isoforms were generously provided by the NCI RAS Initiative, Frederick National Laboratory for Cancer Research, MD, USA, and were cultured in DMEM supplemented with 10% FBS and penicillin (100 units/ml)/streptomycin (100μg/ml). For immunoblotting, cells were lysed in RIPAHi buffer (150 mM NaCl, 50 mM Tris [pH 7.5], 1% NP-40, 0.5% sodium deoxycholic acid, 1% SDS plus Complete protease/phosphatase inhibitor cocktails [Sigma-Aldrich]) and after western blotting, probed with the following primary antibodies: p-ERK Thr202;Tyr204 (E-4), Santa Cruz SC7383; p-EGFR Tyr1068, Cell Signaling 3777; p-ERBB2 Tyr 1248, Millipore 06-229; p-ERBB3 Tyr 1197 Cell Signaling 4561; ERK1/2 Cell Signaling 4695; EGFR Millipore 06-847; ERBB2 Merck OP15L; ERBB3 Millipore 05-390. RAS:RAF binding assays were performed using a commercial RAS activity assay kit (Cytoskeleton) and probed with pan-RAS (AESA02, Cytoskeleton), KRAS (SC-30, Santa Cruz), or HRAS-specific (18295-1-AP, Proteintech) antibodies [41]. Cell propagation and death were analyzed by Incucyte time-lapse video-microscopy in the presence of Sytox green. Cell death measurements were performed after 48 hours of drug treatment and corrected for confluence.
Statistical Analysis
Raw data obtained from quantitative Real Time PCR, FACS, and Incucyte assays were copied into Excel (Microsoft) or Prism (Graphpad) spreadsheets. All mean values, SD, and SEM values of biological replicates were calculated using the calculator function. Graphical representation of such data was also produced in Excel or in Prism. Statistical significance for pairwise data was determined by the Student’s T test. For multiple comparisons, ANOVA was used with a post-hoc Tukey test. * denotes P<0.05; ** denotes P<0.01; *** denotes P<0.001. For Kaplan-Meier plots, log rank P values are presented.
Supplementary Material
One Sentence Summary.
G12 mutant KRAS requires tonic ERBB network activity for initiation and maintenance of lung cancer.
Acknowledgments
We express our gratitude to Peter Adams, Owen Sansom, David Bryant, Martin Eilers, Jennifer O’Neil & Ronan O’Hagan for helpful discussions; to Barbara Bauer, Christine Kramer, Stephen Bell, Derek Miller and the staff of the CRUK Beatson biological services unit for animal husbandry and assistance with animal protocols; to Peter Adams & Nikolay Pchelintsev for help establishing RNA-SEQ; and to all members of the Murphy lab for assistance in manuscript preparation. Ad-SPC-CRE was donated to the Uni. Iowa vector core facility by Anton Berns (NKI).
Funding: Funding was provided by a MINT collaborative grant from Merck Sharpe & Dohme; Deutsche Krebshilfe grant 109220; EC FP7 Marie Curie Actions CIG 618448 “SERPLUC”; British Lung Foundation grants APHD13-5 and CSOBLF RG16-2 (all to DJM). B.K. was supported by EC H2020 Marie Curie actions mobility fellowship 705190 “NuSiCC”. JLQ was funded by MRC programme no. 5TS30.
Footnotes
Author contributions: Experiments were performed and analyzed by B.K.; T.M.; S.N.; A.H.; E. K.; S.L. and S.B.C.. Histology was performed by C.N. and pathology analyzed by J.L.Q. and C.D.. RNA-Seq was performed by W.C. with analysis by A.H.. Statistical guidance was provided by A.H.. Reagents and guidance were provided by K.V. and C.M.. D.J.M. conceived the project, directed experiments and wrote the manuscript. All authors read and approved the manuscript.
Competing interests: The authors declare no conflict of interest.
Materials and data availability: The following RNA-Seq datasets are available from Array Express: E-MTAB-6432; E-MTAB-6483; E-MTAB-6485, & E-MTAB-6486. The Rosa26-lsl-MYC and Hprt-lsl-IRFP mice are available upon request from D.J.M. & K.V., respectively.
References
- 1.Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A, Marcos-Gragera R, et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2) Lancet. 2015;385:977–1010. doi: 10.1016/S0140-6736(14)62038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–51. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351:604–8. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roskoski R., Jr The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacological research. 2014;79:34–74. doi: 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Press MF, Cordon-Cardo C, Slamon DJ. Expression of the HER-2/neu proto-oncogene in normal human adult and fetal tissues. Oncogene. 1990;5:953–62. [PubMed] [Google Scholar]
- 7.Prigent SA, Lemoine NR, Hughes CM, Plowman GD, Selden C, Gullick WJ. Expression of the c-erbB-3 protein in normal human adult and fetal tissues. Oncogene. 1992;7:1273–8. [PubMed] [Google Scholar]
- 8.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13790–5. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, Altman RB, et al. Diversity of gene expression in adenocarcinoma of the lung. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13784–9. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gollamudi M, Nethery D, Liu J, Kern JA. Autocrine activation of ErbB2/ErbB3 receptor complex by NRG-1 in non-small cell lung cancer cell lines. Lung cancer. 2004;43:135–43. doi: 10.1016/j.lungcan.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 11.Chen HY, Liu CH, Chang YH, Yu SL, Ho BC, Hsu CP, Yang TY, Chen KC, Hsu KH, Tseng JS, Hsia JY, et al. EGFR-activating mutations, DNA copy number abundance of ErbB family, and prognosis in lung adenocarcinoma. Oncotarget. 2016;7:9017–25. doi: 10.18632/oncotarget.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Iwanaga K, Choi KC, Wislez M, Raso MG, Wei W, Wistuba II, Kurie JM. Intratumoral epiregulin is a marker of advanced disease in non-small cell lung cancer patients and confers invasive properties on EGFR-mutant cells. Cancer prevention research. 2008;1:201–7. doi: 10.1158/1940-6207.CAPR-08-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sunaga N, Kaira K, Imai H, Shimizu K, Nakano T, Shames DS, Girard L, Soh J, Sato M, Iwasaki Y, Ishizuka T, et al. Oncogenic KRAS-induced epiregulin overexpression contributes to aggressive phenotype and is a promising therapeutic target in non-small-cell lung cancer. Oncogene. 2013;32:4034–42. doi: 10.1038/onc.2012.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–16. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 15.Zhu CQ, da Cunha Santos G, Ding K, Sakurada A, Cutz JC, Liu N, Zhang T, Marrano P, Whitehead M, Squire JA, Kamel-Reid S, et al. Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol. 2008;26:4268–75. doi: 10.1200/JCO.2007.14.8924. [DOI] [PubMed] [Google Scholar]
- 16.de Bruin EC, Cowell C, Warne PH, Jiang M, Saunders RE, Melnick MA, Gettinger S, Walther Z, Wurtz A, Heynen GJ, Heideman DA, et al. Reduced NF1 expression confers resistance to EGFR inhibition in lung cancer. Cancer discovery. 2014;4:606–19. doi: 10.1158/2159-8290.CD-13-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molina-Arcas M, Hancock DC, Sheridan C, Kumar MS, Downward J. Coordinate direct input of both KRAS and IGF1 receptor to activation of PI3 kinase in KRAS-mutant lung cancer. Cancer discovery. 2013;3:548–63. doi: 10.1158/2159-8290.CD-12-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navas C, Hernandez-Porras I, Schuhmacher AJ, Sibilia M, Guerra C, Barbacid M. EGF receptor signaling is essential for k-ras oncogene-driven pancreatic ductal adenocarcinoma. Cancer cell. 2012;22:318–30. doi: 10.1016/j.ccr.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ardito CM, Gruner BM, Takeuchi KK, Lubeseder-Martellato C, Teichmann N, Mazur PK, Delgiorno KE, Carpenter ES, Halbrook CJ, Hall JC, Pal D, et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer cell. 2012;22:304–17. doi: 10.1016/j.ccr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun C, Hobor S, Bertotti A, Zecchin D, Huang S, Galimi F, Cottino F, Prahallad A, Grernrum W, Tzani A, Schlicker A, et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell reports. 2014;7:86–93. doi: 10.1016/j.celrep.2014.02.045. [DOI] [PubMed] [Google Scholar]
- 21.Subramaniam D, He AR, Hwang J, Deeken J, Pishvaian M, Hartley ML, Marshall JL. Irreversible multitargeted ErbB family inhibitors for therapy of lung and breast cancer. Current cancer drug targets. 2015;14:775–93. doi: 10.2174/1568009614666141111104643. [DOI] [PubMed] [Google Scholar]
- 22.Junttila MR, Karnezis AN, Garcia D, Madriles F, Kortlever RM, Rostker F, Brown Swigart L, Pham DM, Seo Y, Evan GI, Martins CP. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature. 2010;468:567–71. doi: 10.1038/nature09526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feldser DM, Kostova KK, Winslow MM, Taylor SE, Cashman C, Whittaker CA, Sanchez-Rivera FJ, Resnick R, Bronson R, Hemann MT, Jacks T. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010;468:572–5. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerr EM, Gaude E, Turrell FK, Frezza C, Martins CP. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature. 2016;531:110–3. doi: 10.1038/nature16967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarkisian CJ, Keister BA, Stairs DB, Boxer RB, Moody SE, Chodosh LA. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nature cell biology. 2007;9:493–505. doi: 10.1038/ncb1567. [DOI] [PubMed] [Google Scholar]
- 26.Moon YW, Rao G, Kim JJ, Shim HS, Park KS, An SS, Kim B, Steeg PS, Sarfaraz S, Changwoo Lee L, Voeller D, et al. LAMC2 enhances the metastatic potential of lung adenocarcinoma. Cell Death Differ. 2015;22:1341–52. doi: 10.1038/cdd.2014.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamata T, Giblett S, Pritchard C. KRAS(G12D) expression in lung-resident myeloid cells promotes pulmonary LCH-like neoplasm sensitive to statin treatment. Blood. 2017;130:514–526. doi: 10.1182/blood-2017-02-770149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou BB, Fridman JS, Liu X, Friedman SM, Newton RC, Scherle PA. ADAM proteases, ErbB pathways and cancer. Expert Opin Investig Drugs. 2005;14:591–606. doi: 10.1517/13543784.14.6.591. [DOI] [PubMed] [Google Scholar]
- 29.Powell CA, Nasser MW, Zhao H, Wochna JC, Zhang X, Shapiro C, Shilo K, Ganju RK. Fatty acid binding protein 5 promotes metastatic potential of triple negative breast cancer cells through enhancing epidermal growth factor receptor stability. Oncotarget. 2015;6:6373–85. doi: 10.18632/oncotarget.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ju JH, Oh S, Lee KM, Yang W, Nam KS, Moon HG, Noh DY, Kim CG, Park G, Park JB, Lee T, et al. Cytokeratin19 induced by HER2/ERK binds and stabilizes HER2 on cell membranes. Cell Death Differ. 2015;22:665–76. doi: 10.1038/cdd.2014.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez-Martin M, Pandiella A. Differential action of small molecule HER kinase inhibitors on receptor heterodimerization: therapeutic implications. International journal of cancer Journal international du cancer. 2012;131:244–52. doi: 10.1002/ijc.26358. [DOI] [PubMed] [Google Scholar]
- 33.Hock AK, Cheung EC, Humpton TJ, Monteverde T, Paulus-Hock V, Lee P, McGhee E, Scopelliti A, Murphy DJ, Strathdee D, Blyth K, et al. Development of an inducible mouse model of iRFP713 to track recombinase activity and tumour development in vivo. Sci Rep. 2017;7 doi: 10.1038/s41598-017-01741-0. 1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cicchini M, Buza EL, Sagal KM, Gudiel AA, Durham AC, Feldser DM. Context-Dependent Effects of Amplified MAPK Signaling during Lung Adenocarcinoma Initiation and Progression. Cell reports. 2017;18:1958–1969. doi: 10.1016/j.celrep.2017.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, Brown-Swigart L, Johnson L, Evan GI. Distinct thresholds govern Myc's biological output in vivo. Cancer cell. 2008;14:447–57. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes & development. 2000;14:2501–14. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grabocka E, Pylayeva-Gupta Y, Jones MJ, Lubkov V, Yemanaberhan E, Taylor L, Jeng HH, Bar-Sagi D. Wild-type H- and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer cell. 2014;25:243–56. doi: 10.1016/j.ccr.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moll H, Pranz K, Musteanu M, Grabner B, Hruschka N, Mohrherr J, Aigner P, Stiedl P, Brcic L, Laszlo V, Schramek D, et al. Afatinib restrains KRAS driven lung tumorigenesis. 2018 doi: 10.1126/scitranslmed.aao2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tebbutt N, Pedersen MW, Johns TG. Targeting the ERBB family in cancer: couples therapy. Nature reviews Cancer. 2013;13:663–73. doi: 10.1038/nrc3559. [DOI] [PubMed] [Google Scholar]
- 40.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & development. 2001;15:3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waters AM, Ozkan-Dagliyan I, Vaseva AV, Fer N, Strathern LA, Hobbs GA, Tessier-Cloutier B, Gillette WK, Bagni R, Whiteley GR, Hartley JL, et al. Evaluation of the selectivity and sensitivity of isoform- and mutation-specific RAS antibodies. Sci Signal. 2017;10:eaao3332. doi: 10.1126/scisignal.aao3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Church DM, Schneider VA, Graves T, Auger K, Cunningham F, Bouk N, Chen HC, Agarwala R, McLaren WM, Ritchie GR, Albracht D, et al. Modernizing reference genome assemblies. PLoS Biol. 2011;9:e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, Watanabe S, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer research. 2012;72:100–11. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.