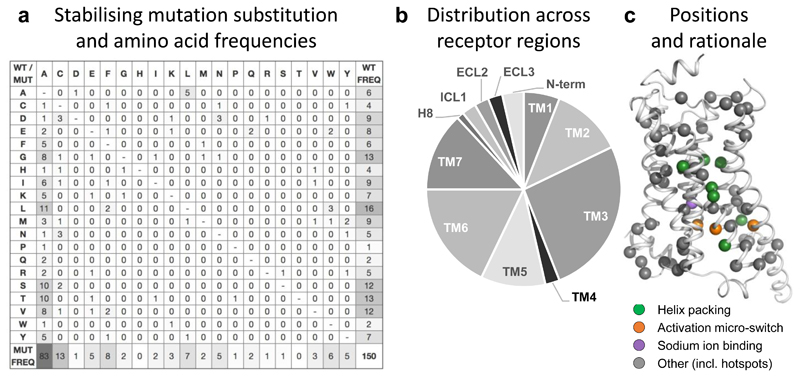
Figure 6.
Substitution frequencies, structural mapping and rationale of stabilising mutations. (a) Amino acid substitution matrix for all 150 stabilising GPCR mutations. The frequencies represent the number of distinct GPCRs and residue positions (merging multiple structures or orthologues) to optimally reflect the transferability across the GPCR superfamily. The different usage of the amino acid alphabet among wild type (rightmost) and mutant (bottom) residues, respectively, is shown with grey-scale. Similarly, grey-scaling is also used to illustrate the success of alanine mutants (leftmost) when substituting mainly similar (small and/or aliphatic hydrophobic) amino acids. (b) Distribution of stabilising mutations (http://gpcrdb.org/construct/stabilisation) across receptor segments, spanning all seven transmembrane helices, the N-terminus, three loops and helix 8. (c) Mapping of all stabilising mutations and their structural rationale onto the β2-adrenoceptor (PDB: 3SN6) using the GPCRdb generic residue indexing34.