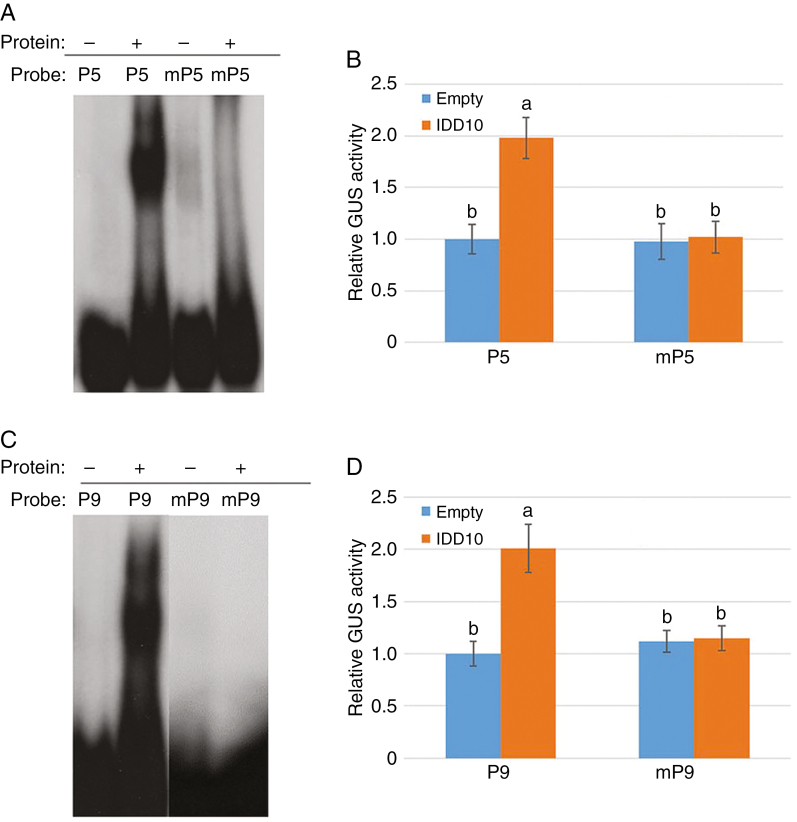
Fig. 5.
Activation of CIPK9 and CIPK14 mediated by binding of IDD10 to IDD-binding motifs. (A) Binding affinities of IDD10 for the native IDD10-binding motif (TTTGTCG; P5) from the CIPK9 promoter and a mutated motif (TTTTTTT; mP5). (B) Activation of the native CIPK9 promoter (P5) and the mutated promoter (mP5) in protoplasts co-transfected with IDD10. (C) Binding affinities of IDD10 for the native IDD10-binding motif (AAACAGG; P9) from the CIPK14 promoter and a mutated motif (AAAAAAA; mP9). (D) Activation of the native CIPK14 promoter (P9) and the mutated promoter (mP9) in protoplasts co-transfected with IDD10. Binding affinities (A and C) of IDD10 for native (P5 and P9) and mutated (mP5 and mP9) putative IDD10-binding motifs were determined using EMSA. Transient expression assays (B and D) were performed in arabidopsis protoplasts co-transfected with IDD10 and vectors expressing GUS under the control of a 1.5 kb promoter from CIPK9 (B) or CIPK14 (D) containing either native or mutated IDD-binding motif sequences; co-transfection with empty vector was used as a control. Luciferase expression driven by the 35S promoter was used as an internal control for normalizing GUS expression data. Data in (B) and (D) are means ± s.e. (n = 3); different letters indicate significant differences between results (P < 0.05).