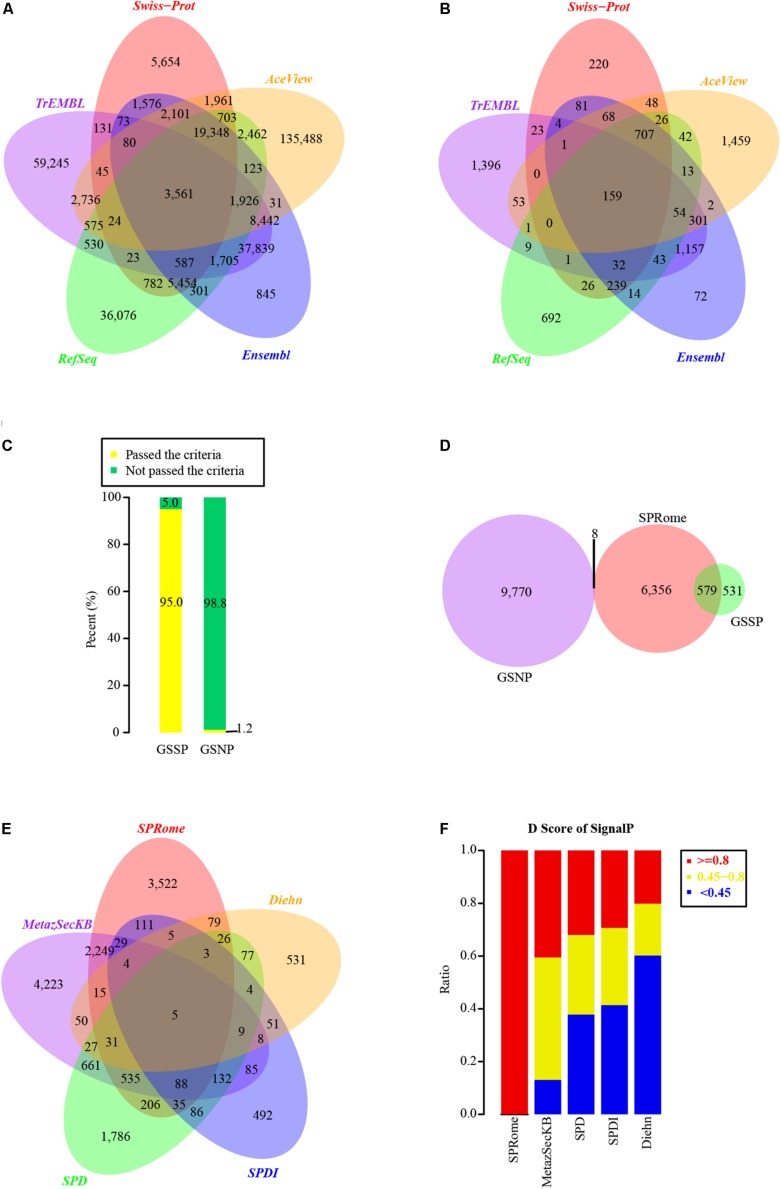
FIGURE 1.
Identification and evaluation of SPs based on comprehensive human protein set. (A) Venn graph of human proteins used for SP identification in different databases. A total of 330,427 non-redundant proteins were integrated from Swiss-Prot, TrEMBL, RefSeq, Ensembl, and AceView databases. (B) The distribution of SPs identified in Swiss-Prot (1,635), TrEMBL (3,234), RefSeq (2,058), Ensembl (2,947), and AceView (2,934) databases. (C) The percentages of proteins passed and not passed the default SignalP cutoff (D-score >0.45) in GSSP and GSNP. (D) Comparison of our identified SPs with GSSP and GSNP. (E) Comparison of our identified SPs with other four known SP sets. (F) Distribution of SignalP D-scores in our identified SPs and other four SP sets.