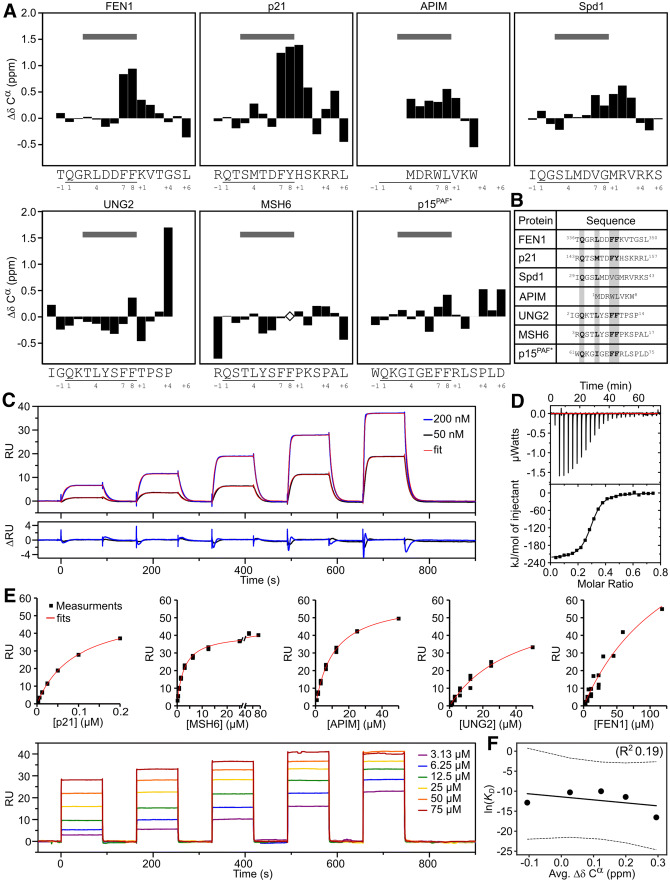
Fig. 2.
Preformed structure in disordered PCNA ligand motifs does not correlate to affinity. a Secondary Cα-chemical shifts (ΔδCα) per residue of unbound PCNA target peptides. The sequences of the peptides and motif numbering (from − 2 to + 6) are shown below each plot, with PIP-motif-positions underlined. The positions of the 310-helix-forming residues of p21 in complex with PCNA (position 3–8) are marked at the top with a gray bar. Open diamonds indicate lack of assignment. For Spd1 and p15PAF the assignments are from full-length proteins. The MSH6, FEN1, p21, and APIM peptides were N-terminally acetylated and C-terminally amidated. *From BMRB entry 1933 [50]. b Alignment of peptide sequences with numbering from the full-length proteins. PIP box specific residues are highlighted in gray. c Binding of p21 to human PCNA by SPR. Sensorgrams were obtained by injecting a series of p21 concentrations over PCNA captured by the immobilized Anti-His6 antibody. The p21 concentrations were five serial twofold dilutions, injected in the order of increasing concentrations, with a final concentration of 50 nM (black) or 200 nM (blue). Non-linear regression fits are shown in red with residuals below. d Binding of p21 to human PCNA by ITC. In the representative ITC experiment PCNA was injected into the p21 peptide. The upper portion shows baseline-corrected raw data from the titration, and the low portion shows the normalized integrated binding isotherms with the fitted binding curves assuming a single set of equivalent binding sites. e Equilibrium SPR binding analyses of PCNA ligands (p21, MSH6, APIM, UNG2, and FEN1). The lower part shows a representative binding series of MSH6 to PCNA, with the concentration of the final injection indicated. The sensorgrams were obtained by injecting the respective peptide over PCNA captured by the immobilized anti-His6 antibody. The BIAevaluation software was used to extract the KDs, fits shown in red. f Correlation between average SCS of the PCNA ligands in a and ln(KD) (KDs in M as measured by SPR in c and e)