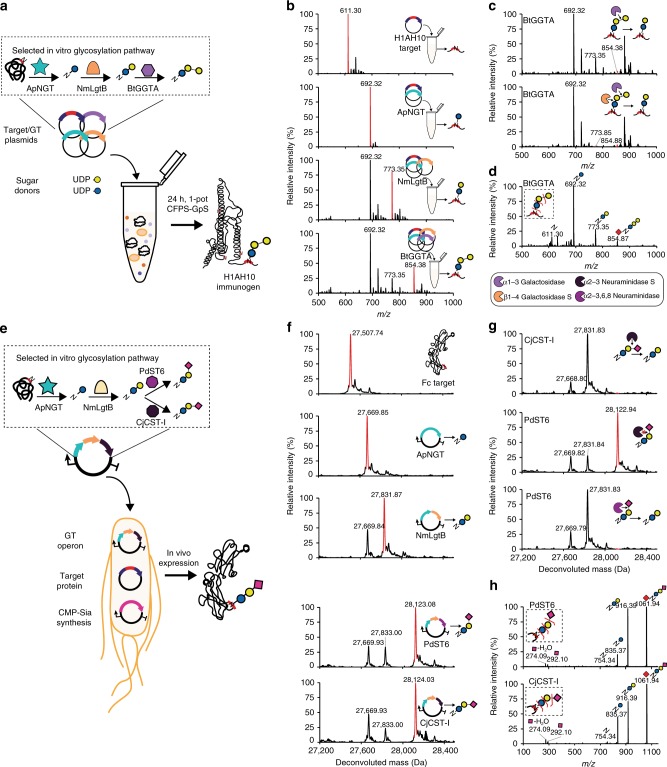
Fig. 4.
Design of biosynthetic pathways for cell-free and bacterial production platforms. a One-pot CFPS-GpS for synthesis of H1HA10 protein vaccine modified with αGal glycan. Plasmids encoding the target protein and biosynthetic pathway GTs discovered by GlycoPRIME screening were combined with appropriate activated sugar donors in a CFPS-GpS reaction. b Trypsinized glycopeptide MS spectra, c exoglycosidase digestions of glycopeptide, and d MS/MS glycopeptide fragmentation spectra from H1HA10 purified from IVG reactions containing equimolar amounts of each indicated plasmid encoding H1HA10, ApNGT, NmLgtB, and BtGGTA and 2.5 mM of UDP-Glc and UDP-Gal (see “Methods”). All reactions contained 10 nM total plasmid concentration and were incubated for 24 h at 30 °C. The glycopeptide contains one engineered acceptor sequence located at the N-terminus of H1HA10. Observed masses and mass shifts in (b–d) spectra are consistent with modification of the H1HA10 peptide with N-linked Glc by ApNGT, lactose (Galβ1-4Glc-Asn) by ApNGT and NmLgtB, or αGal epitope (Galα1-3Galβ1-4Glc-Asn) by ApNGT, NmLgtB, and BtGGTA. e Design of cytoplasmic glycosylation systems to produce sialylated IgG Fc in E. coli. Three plasmids containing NmNeuA (CMP-Sia synthesis), IgG Fc engineered with an optimized acceptor sequence (target protein), and biosynthetic pathways discovered using GlycoPRIME (GT operon). f Deconvoluted intact glycoprotein MS spectra, g exoglycosidase digestions of intact glycoprotein, and h MS/MS glycopeptide fragmentation spectra from Fc-6 purified from E. coli cultures supplemented with sialic acid, IPTG, and arabinose and incubated at 25 °C overnight (see “Methods”). The last GT in all glycosylation pathways is indicated. MS spectra were acquired from full elution areas of all detected glycosylated and aglycosylated protein or peptide species and are representative of n = 3 CFPS-GpS or E. coli cultures. MS/MS spectra acquired by pseudo multiple reaction monitoring (MRM) fragmentation at theoretical glycopeptide masses (red diamonds) corresponding to detected intact glycopeptide or protein MS peaks using 30 eV collisional energy. Deconvoluted spectra collected from m/z 100–2000 into 27,000–29,000 Da using Compass Data Analysis maximum entropy method. See Supplementary Tables 5–7 for theoretical masses