Fig. 2.
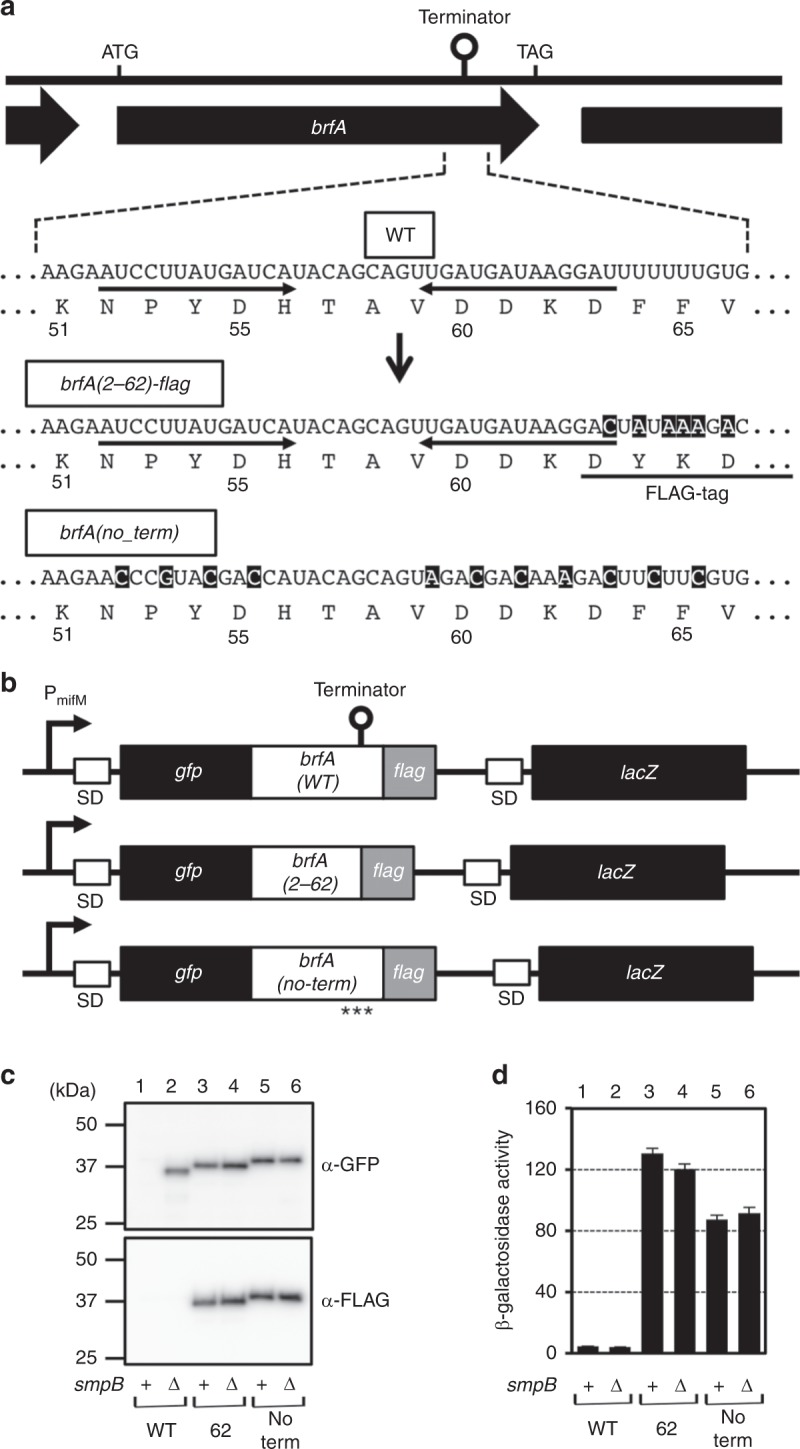
The cellular abundance of BrfA is negatively regulated by trans-translation. a A schematic representation of the brfA open reading frame, showing the intrinsic transcriptional terminator sequence within the coding region (indicated by “terminator” at the top and the wild type (WT) enlarged view below). Shown also are the corresponding sequences of the gfp-brfA(2–62)-flag and gfp-brfA(no_term)-flag constructs (see b), in which nucleotide substitutions and the FLAG tag are indicated by reverse and underline, respectively. b Schematic representations of the gfp-brfA-flag constructs with wild-type terminator sequence (gfp-brfA(2–71)-flag) and its derivatives with defective terminator signals. These gfp-brfA derivatives were placed under the constitutive mifM promoter. The lacZ gene is also placed downstream of the gfp-brfA derivatives. “SD” and asterisks indicate Shine-Dalgarno sequence and synonymous mutations, respectively. c Cellular accumulation of the products of the wild-type construct (lanes 1, 2) as well as the brfA(2–62) (lanes 3, 4) and the brfA(no_term) (lanes 5, 6) constructs. They were expressed in the smpB+ (odd numbers) or the ΔsmpB (even numbers) strains and analyzed by anti-GFP (upper) or anti-FLAG (lower) immunoblotting. d β-Galactosidase activities (mean ± s.d., n = 3) of the cells horbaring lacZ at the downstream of wild type (columns 1, 2), the brfA(2–62) (columns 3, 4), or the brfA(no_term) (columns 5, 6) derivatives of the gfp-brfA in the presence (odd numbers) or absence (even numbers) of smpB. “s.d.” indicates standard deviation. Source data are provided as a Source Data file.
