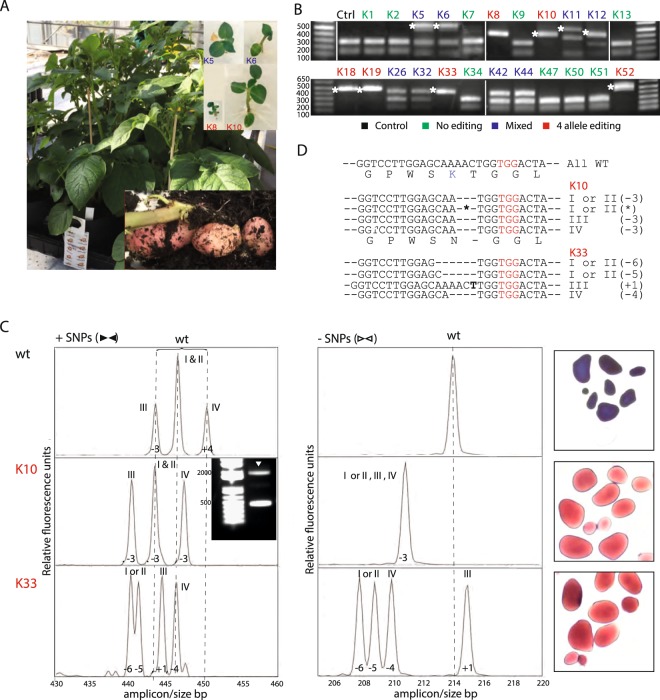
Figure 2.
Amylopectin only potato: Geno- and phenotypic analysis of GBSS loss of function ex-plants. (A) Regenerated ex-plants from tissue culture with resulting potato tubers.(B) In total, 23 randomly selected shoots/ex-plants were screened to identify indels using the BsrI restriction enzyme digestion. K8, K10, K18, K19, K33 & K52 appeared to have editing in all four alleles (fully BsrI resistant), K11, K12, K26, K32 & K42 displayed a mixture of WT type and edited alleles, while K1, K2, K7, K9, K13, K34, K47, K50 & K51 appeared to be un-edited. * denotes insertions. K5 and K6 appear at a first glance to be mixed, but sequence analysis revealed that the BsrI digestion bands were derived from plasmid insertions comprising the gRNA1 sequence, which reintroduced the BsrI site and that K5 and K6 in fact had full allelic editing. (C) WT and the full allelic edited ex-plants, K10 and K33, were corroborated by IDAA of the larger (+SNP) region and the inner smaller (-SNP) region without length SNPs (Fig. 1C). WT peak positions are indicated by dotted lines. Lugol staining was used to analyse tuber starch from greenhouse grown plants (Supplementary Fig. 6). The presence of amylose gives rise to the dark blue colour (WT) tubers, while amylose free/’amylopectin only’ yields the red-brownish color. Additional shoots/ex-plants selected for IDAA analysis of which a subset was propagated to set tubers were also stained with lugol29 (Supplementary Fig. 7). (D) Sequence analysis confirmed that both K10 and K33 were full allelic edited. K10 had a 3 bp deletion in allele III, IV and in one of the alleles I or II and a 990 bp insertion (denoted ‘*’ left to the sequence, see also arrow on gel insertion (Fig. 2C)) in the other as also evident in the IDAA analysis displaying 1:1:1 ratio of the three chromatographic peaks. K33 had 6 bp and 5 bp deletions in allele I and II, a 4 bp deletion in allele IV and a 1 bp insertion in allele III.