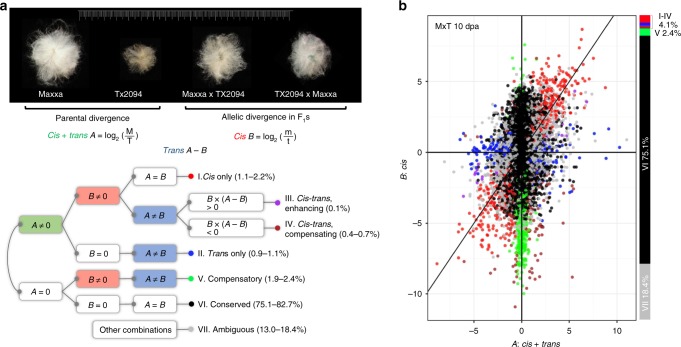
Fig. 1.
Parental and F1 hybrid expression data enable the ASE study of G. hirsutum domestication. a Using parental and F1 allelic expression divergence between wild and domesticated cottons, genes were assigned into one of seven regulatory categories representing combinations of cis and trans regulatory effects. Briefly, differential expression of any given gene in the parents reflects both cis and trans divergence (A), whereas the expression of the same gene in the common trans environment of the F1 hybrid reflects cis regulatory divergence (B) between parental alleles. While trans divergence cannot be directly measured, it can be inferred via the difference in A − B. See methods for additional description. Next to each category, the percentage range of genes was obtained from four sample conditions (M × T and T × M hybrids each at 10 and 20 dpa). b Taking the 10 dpa sample of the F1 hybrid M × T as an example, the scatter plot of cis regulatory divergence (y-axis) vs. parental expression divergence (x-axis) is shown for all seven categories of genes. Category I–IV together only account for 4.1% of 27,816 genes, with the majority of genes assigned to categories of conserved (VI–75.1%), ambiguous (VII–18.4%), or compensatory regulation (V–2.4%). The source data underlying Fig. 1b are provided as a Source Data file.