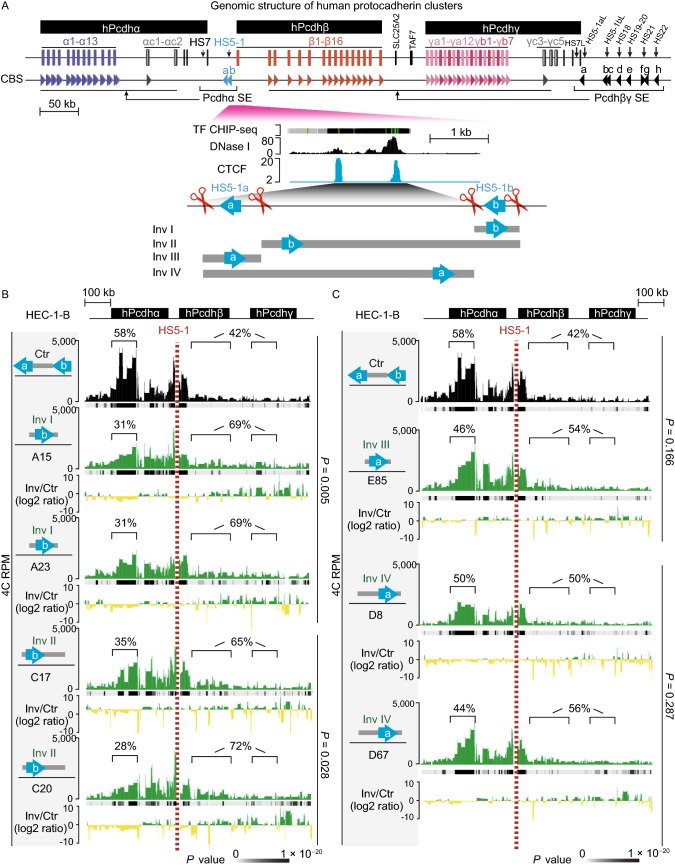
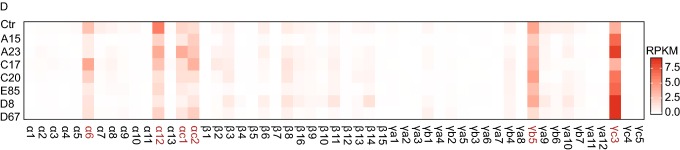
Figure 1.
Genetic dissection of thePcdhα HS5-1enhancer architecture by CRISPR DNA-fragment editing. (A) Schematics of the three human Pcdh gene clusters (hPcdh α, β, and γ) and the inversion experiments mediated by CRISPR DNA-fragment editing. The orientation of CBS arrays is indicated by horizontal arrowheads. The DNase I hypersensitive sites within super-enhancers are indicated by vertical arrows. The HS5-1 enhancer and its various CRISPR inversions are highlighted below. Two tandem CBSs in a reverse orientation (HS5-1a and HS5-1b) at the editing sites are also highlighted. SLC25A2 and TAF7 are two non-Pcdh genes. CBS, CTCF-binding site; HS, hypersensitive site; SE, super-enhancer; TF, transcription factor. (B) Shown are 4C chromatin-interaction profiles in the wild-type control (Ctr) and CRISPR cell lines with inversion of the CBS HS5-1b only (Inv I) or the HS5-1b combined with the middle region (Inv II) using the HS5-1 enhancer as a viewpoint. The significance of interactions (P value) is shown under the read’s density. The log2 ratios between each inversion cell clone and wild-type cells are also indicated. (C) Shown are 4C chromatin-interaction profiles in wild-type control and CRISPR cell lines with inversion of the CBS HS5-1a only (Inv III) or the HS5-1a combined with the middle region (Inv IV) using the HS5-1 enhancer as a viewpoint. (D) Heatmap shows the alteration of expression patterns of members of the three Pcdh gene clusters resulting from CRISPR inversion of different enhancer and insulator (CBS) elements by RNA-seq experiments