Figure 1: Nitrogen is not essential for the initial expansion of metabolism.
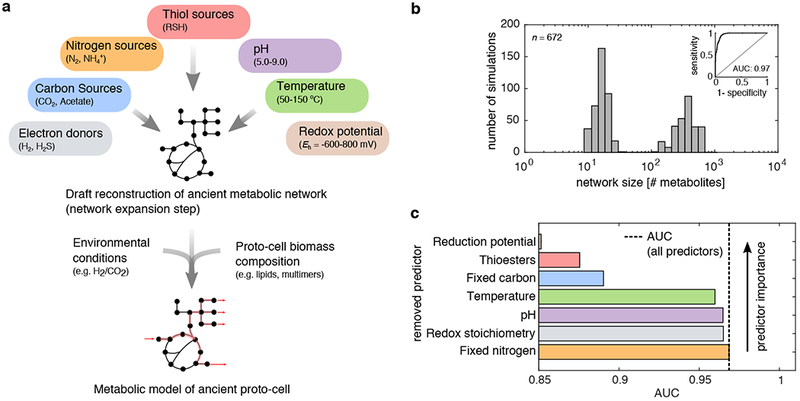
(a) A network expansion algorithm (top) was used to simulate the early expansion of metabolism under 672 scenarios, systematically varying the availability of reductants in the environment, pH, carbon sources, the presence of thiols, temperature and the availability of ammonia. This process is subject to local thermodynamic feasibility constraints, i.e. allows new reactions to occur only if they are individually thermodynamically feasible (see Methods). For a subset of networks obtained from network expansion, we implemented detailed stoichiometric model simulations, using flux balance analysis (bottom), in which we implemented global thermodynamic feasibility constraints (see also Methods and Fig. 4). (b) A histogram of network sizes (x-axis, number of metabolites) revealed that 43 % (288/672) of the scenarios resulted in a bimodal distribution, where expansion occurred beyond 100 metabolites. (inset) A logistic regression classifier was constructed to predict whether a geochemical scenario resulted in a network that exceeded 100 metabolites, and a receiver operating curve (ROC) was plotted. The trained classifier resulted in an area under the curve (AUC) of 0.97 and leave-one out cross-validation accuracy of 0.89. (c) Models were trained without information on specific geochemical variables (y-axis, ranked by predictor importance), and the ensuing AUC was plotted as a bar-chart (x-axis), revealing that knowledge of the availability of fixed nitrogen offers no information on whether networks expanded.
