Figure 2: Primitive redox systems and reaction classes constrain network expansion from CO2.
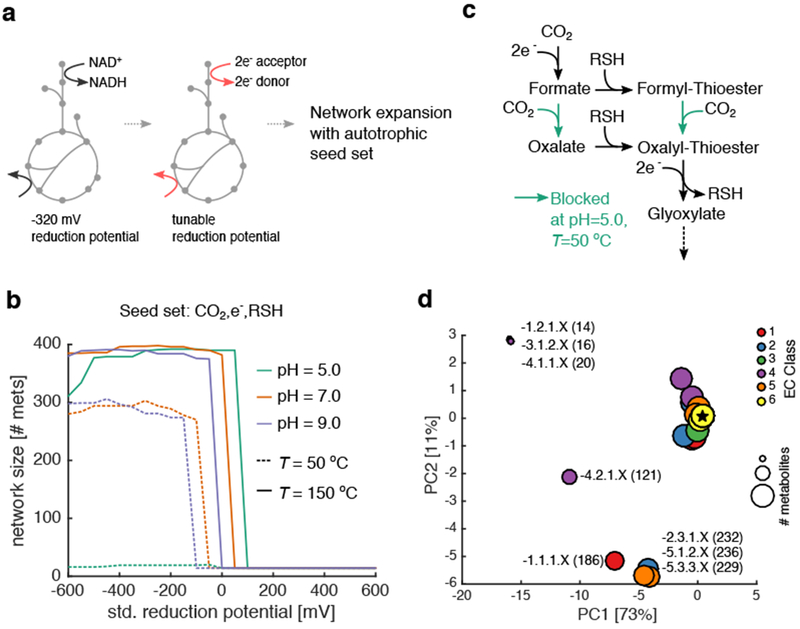
(a) Redox coenzymes (NAD, NADP, and FAD) were substituted with an arbitrary electron donor/acceptor at a fixed reduction potential. (b) We performed thermodynamic network expansion in acidic (pH 5), neutral (pH 7) and alkaline (pH 9) conditions at two temperatures (T =50 and 150 °C), using a two-electron redox couple at a fixed potential (x-axis) as a substitute for NAD(P)/FAD coupling in extant metabolic reactions (see Methods). We plotted the final network size across all pH and temperatures with no fixed carbon sources (e.g. only CO2) and thiols. Notably, for these simulations, we used a base seed set of: H2, H2S, H2O, HCO3−, H+ and CO2. In general, network size is increased at higher temperatures, consistent with Weiss et. al 13. (c) Analysis of the expanding networks revealed a critical set of reactions blocked at pH 5 and T=50 °C, preventing the production of oxalyl-thioesters and subsequently glyoxylate. These reactions belong to Enzyme Commission (EC) class 4.1.1.X (d) We performed network expansion after removing groups of reactions based on EC codes, and performed principal component analysis (PCA) on the ensuing networks. The color represents the EC code removed from the original network, while the size corresponds to the final number of metabolites in the expanded network. Points are labeled by E.C. class, and the number of metabolites in the perturbed networks are provided in parentheses. The star represents the location of the unperturbed network.
