Figure 4: Constraint-based modeling of plausible ancient proto-cells.
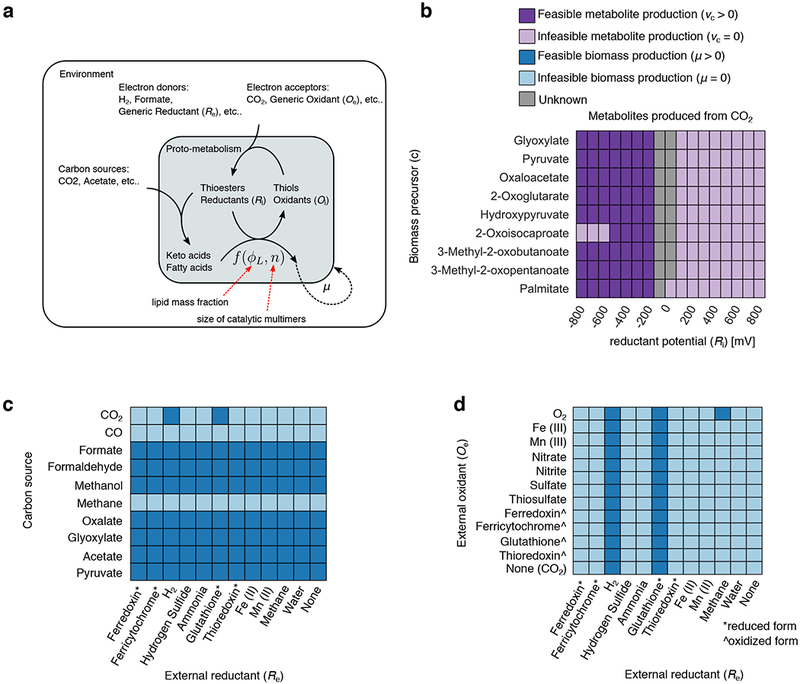
(a) We constructed a metabolic model of a plausible ancient proto-cell and used thermodynamic metabolic flux analysis53 to simulate the feasibility of steady state growth under a variety of environmental conditions. The metabolic model was constructed using internally-generated reductants (Ri) and thioesters that fueled biomass formation. The biomass composition was specified as variable fractions of fatty acids, and polymerized hydroxy-acids from keto-acid precursors (see Methods). In this model, the internal redox coenzyme was assumed to be at a single fixed standard reduction potential, and the production of biomass was fueled by the hydrolysis of acetyl-thioesters. We parameterized the biomass composition using a two-parameter model (see Methods), with the mass fraction of lipids in the proto-cell set to ϕL = 0.1, and the average size of a catalytic multimer, n = 10. (b) We computed fluxes from CO2 to each biomass precursor (y-axis) using a variety of internally generated reductants at various reduction potentials (x-axis), and show the conditions that led to feasible (dark purple) and infeasible (light purple) flux. Note that some cases did not converge to a solution within the allocated maximal CPU time, likely due to numerical issues, and were thus classified as “unknown” (grey). The production of all biomass precursors was feasible if the redox system was between −500 and −200 mV. (c) We next simulated growth on a variety of simple carbon sources (y-axis) and external electron donors (x-axis) and show environments supporting non-zero growth (light blue) or no growth (dark blue). Interestingly, H2 and glutathione were the only reductants capable of supporting fully autotrophic growth on CO2. Furthermore, CO and Methane could not support growth in this model, while the other one-carbon sources like methanol, formate and formaldehyde could support biomass growth. (d) We next simulated autotrophic growth using both an oxidant (y-axis) and reductant (x-axis) and show environments supporting non-zero growth (dark blue) or no growth (light blue). Feasible growth was entirely dependent on the reductant, rather than the oxidant, except for the case with methane as the electron donor and oxygen as the electron acceptor. For models in (c) and (d), the internal redox coenzyme was assumed to be disulfide/dithiol at a standard reduction potential of −220 mV.
