Figure 1.
Characterization of Bovine circTTN
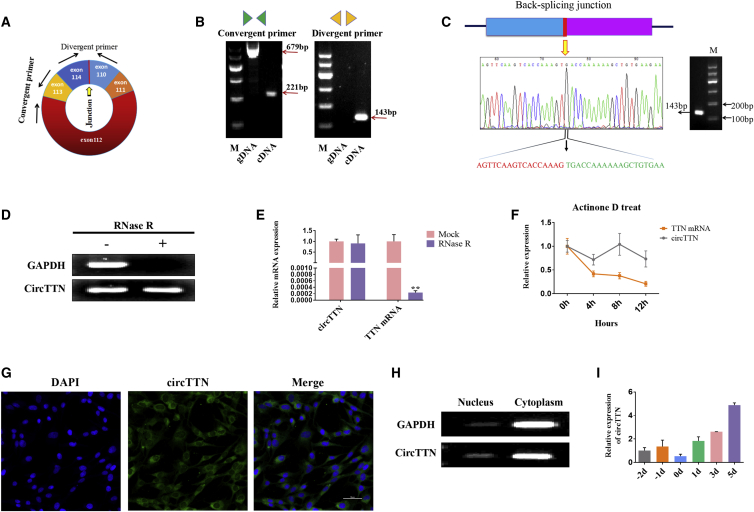
(A) Schematic view illustrating the design of primers for circTTN. A divergent primer was also used in quantitative real-time PCR. (B) A convergent primer and divergent primer were used to confirm the circular nature of circTTN. (C) The circular junction of circTTN was identified by using a divergent primer on Sanger sequencing. (D) RNase R detected the presence of circTTN. (E) The expression of circTTN and TTN mRNA in myoblasts treated with RNase R was determined by quantitative real-time PCR. (F) Quantitative real-time PCR for the abundance of circTTN and TTN mRNA in bovine primary myoblasts treated with Actinomycin D at the indicated time points. (G) RNA-FISH assay was performed to determine circTTN subcellular localization. Blue indicates nuclei stained with DAPI; green indicates the RNA probe that recognizes circTTN. Scale bar, 50 μm. (H) The expression of circTTN in the cytoplasm and nuclear was detected by semiquantitative PCR. (I) The expression of circTTN in myoblasts differentiated for −2, −1, 0, 1, 3, and 5 days is shown. Data are presented as means ± SEM for three individuals.