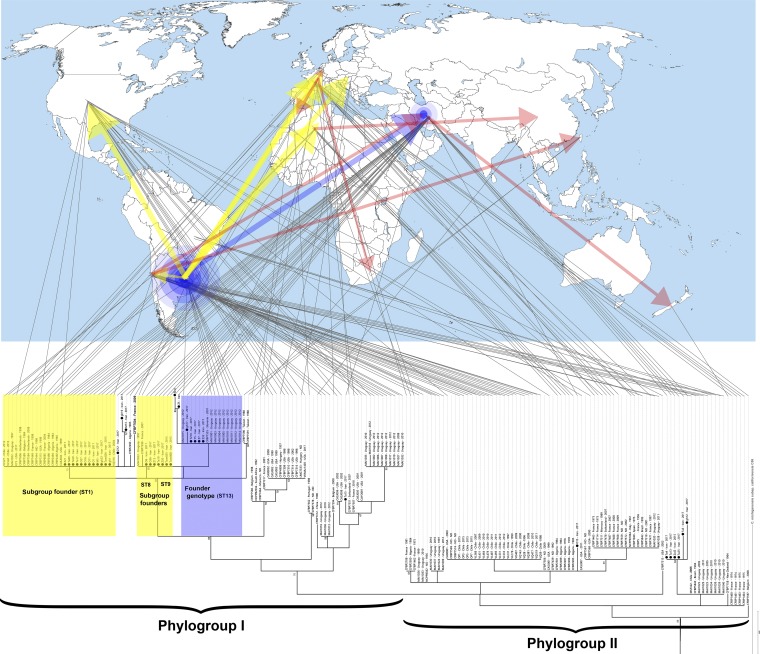
FIG 2.
Phylogeny of Clavibacter michiganensis subsp. michiganensis strains isolated in this study among the worldwide population of the pathogen based on the concatenated sequences of four housekeeping genes (i.e., atpD, gyrB, ppk, and recA). Clavibacter michiganensis subsp. californiensis was used as an outgroup to root the tree. The bar presents numbers of substitutions per site. Gray lines linking the phylogenetic tree to the world map indicate geographic origin of a given strain. Strains isolated in Iran are indicated by black circles. Worldwide C. michiganensis subsp. michiganensis strains were divided into two phylogroups (I and II) based on the date of isolation. Blue highlighting on the tree indicates the founder sequence type (ST13), while the yellow highlighting shows the subgroup founders. Blue concentric circles on the map indicate the predicted center of diversity of the pathogen. Yellow arrows show the hypothetical transmission route of subgroup founders, while red arrows represent the subsequent transmission routes of the pathogen. The source map is from https://commons.wikimedia.org/wiki/File:A_large_blank_world_map_with_oceans_marked_in_blue.PNG.