Fig. 5.
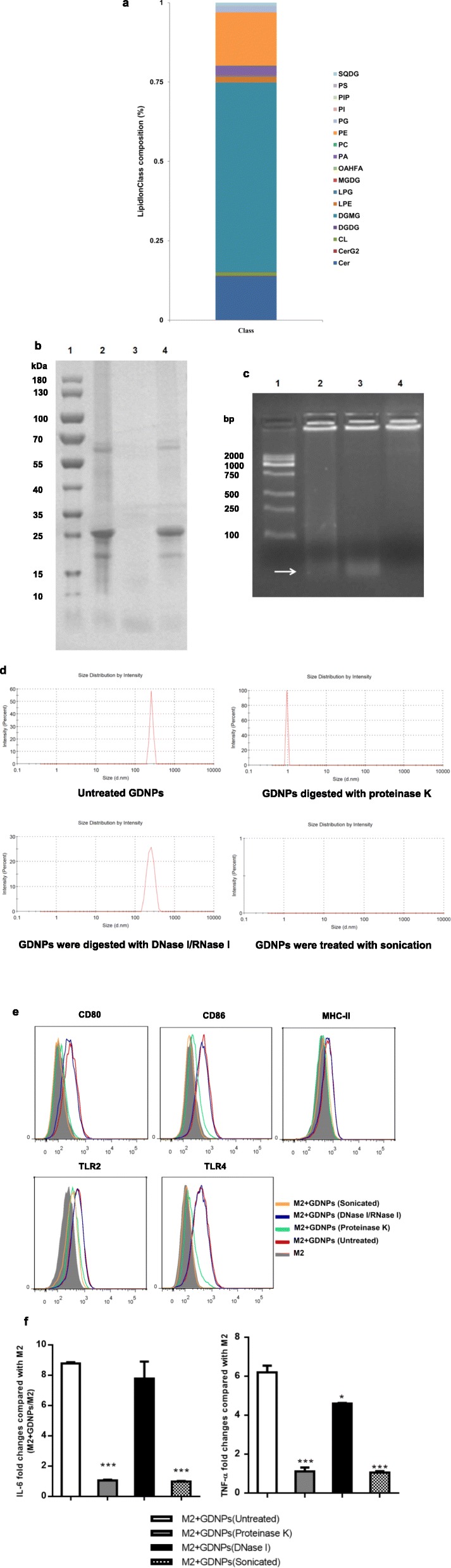
Analysis of the GDNPs components involved in macrophage polarization. a The percentages of lipid species in GDNPs. b GDNPs were digested with proteinase K. The proteins in GDNPs were analyzed via 12% SDS polyacrylamide gel electrophoresis. (1.Markers, 2.GDNPs, 3.GDNPs digested with proteinase K, 4. GDNPs digested with DNase I/RNase I). c GDNPs were digested with DNase I/RNase I. The nucleic acids in GDNPs were analyzed via agarose gel electrophoresis. (1.Markers, 2.GDNPs, 3.GDNPs digested with proteinase K, 4. GDNPs digested with DNase I/RNase I). d Particle size of the GDNPs was treated with different reagents. e GDNPs (10 μg/ml) subjected to the indicated treatments were co-incubated with M2-like macrophages for 48 h. Macrophages were collected and stained with antibodies against the indicated surface markers. The expression of surface markers on macrophages was analysed by flow cytometry (grey shaded histograms indicate complete medium; red line indicates exposure to untreated GDNPs; green line indicates exposure to GDNPs treated with proteinase K; blue line indicates exposure to GDNPs treated with DNase I and RNase I; orange line indicates exposure to sonicated GDNPs). f M2-like macrophages were incubated with GDNPs subjected to the indicated treatments for 48 h; IL-6 and TNF-α in the supernatants were measured by ELISAs. The results represent three independent experiments as the mean ± SEM. **P < 0.01, ***P < 0.001 compared with M2 + GDNPs (untreated); evaluated using Student’s t test (f)
