Abstract
Objective:
The objective of the present study was to evaluate heterozygosis in cattle population, and to characterize White Fulani breed by identifying DNA markers considering microsatellites.
Materials and Methods:
A total of 41 cattle were randomly selected and used for sample (wool) collection for the characterization and identification of phenotypic traits of cattle in Nigeria. The DNA samples from the samples were prepared. Twelve microsatellite primers were used for the microsatellite analysis in the genomic DNA of cattle. The reinforced products were analyzed to determine polymorphic alleles and their frequencies.
Results:
White Fulani is characterized by a high degree of genetic diversity. The microsatellites have multiple alleles and may show heterozygosity frequencies of at least 70%. White Fulani cows and their F1 descendants form a common cluster, to which the bulls of the Kuru and Red Boro breeds are adjacent. There is a clear differentiation of purebred populations of Tajik zebu-like cattle (Q = 98.7%) and a significant proportion of white Fulani (Q = 81.8%) from Nigeria. The microsatellite analysis of zebu of Nigeria allowed identifying a total of 80 alleles. In the KURU and PAX-KR-BOR rocks, 17 and 19 alleles were identified, respectively. In F1, 51 alleles were detected.
Conclusion:
White Fulani cattle are characterized by a high degree of genetic diversities. This makes it a highly informative source in genetic analysis. The results can be applied in dealing with the conservation and sustainable applications of genetic resources in the Nigerian cattle population.
Keywords: Genetic, microsatellite loci, phenotypic, polymorphic alleles
Introduction
The original habitat of White Fulani breed is in Northern and Southern Nigeria, and the Northeastern part of Cameroon, associated with the Fulani and House lemens; gradually spread to the south of Chad, and western Sudan (where they are called Fellata, and red Fulani) [1]. In Nigeria, this breed accounts for about half of the Nigerian total national herds, and about 95% of them are pure breed of Fulani [1–3].
Fulani is characterized by long (80–105 cm) and lyre horns, white fur, interspersed with black, sometimes with red marks on the ears, legs, and sides. This cattle breed has great potential as dual-use (milk and beef) [4]. Fulani breeds include Nigerian Fulani (Gyelly, Diali), Fulani of Senegal (Fowloule, Hump), Sudanese Fulani (Zebu Paul Sudanais), and White Fulani (Bunai, Yakanai) [5]. Zebu Fulani found in the territory, located to the western regions of Senegal to areas located to the east of Lake Chad [6]. In terms of weight, meat, and milk productivity (efficacy), Fulani of the listed breeds clearly differed from each other; White Fulani cattle is an important beef cattle breed reared in the Sahel region of Africa [4].
White Fulani (synonymous with Akou Republic of Cameroon, Bunaji, White Bororo, White Kano, and Yakanji) is located in northern Nigeria, and in the Federal Republic of Cameroon in a climatic environment, which is characteristic of semi-arid, where it is called Akou White Fulani (local name Yakanji). In terms of phenotype, the coat is usually white with black dots, but some animals have a black suit with blue, red, or white spots. The skin is loose with pigmented soft hair. The ears are erected. The horns are medium or long and lyre-shaped. White Fulani is commonly used for meat, milk, and draught [4, 6–8, 9, 11].
The single nucleotide polymorphism (SNP) markers have gained the excessive reputation, even though they are only bi-allelic co-dominant markers [12]. Currently, two different SNP marker sets were identified [13, 14] for animal identification and parentage testing in American and European beef cattle populations. To achieve productivity gains (high milk production); White Fulani breed is silently being upgraded via artificial insemination with the semen of Bos taurus breeds, especially Friesian [2]. As a result, crossbred Bunaji is developed, which is considered as a threat to the original Bunaji resources [15].
In spite of the availability and importance of these cattle breeds (Bunaji), very little work has been carried out to characterize them phenotypically. Characterizing them will give a good knowledge of the diversity and relationship that exists between them. It will enable quick identification and their productive performance [15]. The importance of maintaining such diversity had been emphasized in several reports [7, 16, 17]. A cattle breed of Kuru Lake Chad (buduma) is well developed only on the rich salts of potassium coasts; a characteristic feature is the giant clavate horns, having 33-35 cm in circumference. high-legged animals, late-ripening, with flat ribs, height at the withers of cows up to 138 cm, live weight up to 400 kg. Milk productivity is low and the bone content in the carcass is relatively high [16]. The objective of the present study was to evaluate heterozygosis of White Fulani cattle population by identifying DNA markers considering microsatellites, and the genetic characterization was done comparing with other cattle breeds.
Materials and Methods
Animals were handled and experiments were conducted in accordance with regulations of the People’s Friendship University of Russia and its ethical rules.
Studies were carried out in the Laboratory of Molecular Genetics of Animals at the Center for Biotechnology and Molecular Diagnostics of the VIZH Russian Agricultural Academy in Molecular Genetic Studies. The study was conducted on samples of zebu DNA 3 breeds of different directions of productivity and hybrids of the first generation between them F1. Local zebu cattle of Tajikistan were taken as the comparison group [7].
The DNA extraction material was used to sample the zebu wool from which the DNA bank of three different zebu breeds and first-generation crossbred animals was formed. Immediately after collection, the wool samples were stored at room temperature until they were delivered to the laboratory and stored at +4°C in the DNA Bank of the Center for Biotechnology and Molecular Diagnostics of Animals of the Russian Agricultural Academy. DNA isolation was performed using sodium perchlorate and using the DIAtomTM DNA Prep100 reagent kit (Laboratory Isogene, Russia) for DNA extraction according to the recommendations of the manufacturer. DNA analysis and polymerase chain reaction (PCR) were performed, as per the methods described by Brežná and Piknová [18].
The set of markers for analysis included the following single-probe loci: 12-plex PCR—TGLA53, TGLA122, TGLA126, TGLA227, INRA023, ETH10, ETH3, ETH225, BM1818, BM1824, BM2113, and SPS115.
Molecular characterization was performed on the zebu-like cattle of the following sex-age groups: White Fulani cows, Red Bororo-Rahaji, and Kuru bulls, as well as the first generation interbreeding between them F1. For the purpose of comparative assessment of genetic diversity and population-genetic parameters, the analysis of microsatellite profiles of the studied cattle breeds in Nigeria was carried out in comparison with other breeds of livestock bred in Nigeria, and Zebuvid livestock of Tajikistan.
The data were analyzed used the general Linear Model. Breed and gender were included as fixed variables and the length of body and other characterization were proposed as continuous variables. The significance of variables was separated due to the least significant difference. The for-more analysis is a resulted in multiple regression models. SAS software was applied for statistical analysis.
Results and Discussion
The microsatellite analysis of the whole studied sample of zebu of Nigeria allowed identifying a total of 80 alleles, with the maximum allelic diversity being detected in the BON-B-FUL breed—60 alleles. In the KURU and PAX-KR-BOR rocks, 17 and 19 alleles were identified, respectively. In F1, 51 alleles were detected. The decrease in the total number of detected alleles is a consequence of the small size of the sample being analyzed. The results of the analysis of the total and effective number of alleles in the locus and the Shannon index, as well as the average number of alleles per locus in the pedigree aspect, are summarized in Table 1. As mentioned in Table 1, the number of alleles in the MS locus in groups of cattle of various breeds varied from two in the ETH225 and BM1824 loci to nine in the SPS115 and INRA23 loci and all in the BON-B-FUL breed.
Table 1. The actual and expected degree of heterozygosity in the zebu breeds studied.
| Population | Degree of heterozygosity | Difference (Ho – He) | Fis | |
|---|---|---|---|---|
| Actual (Ho) | Expected (He) | |||
| BON-B-FUL | 0.622 ± 0.073 | 0.686 ± 0.046 | −0.064 | −0.004 ± 0.183 |
| KURU | 0.700 ± 0.153 | 0.350 ± 0.076 | 0.350 | −1.000 ± 0.000 |
| RAX-KR-BOR | 0.900 ± 0.100 | 0.450 ± 0.050 | 0.450 | −1.000 ± 0.000 |
| F1 | 0.710 ± 0.071 | 0.723 ± 0.025 | −0.013 | 0.019 ± 0.090 |
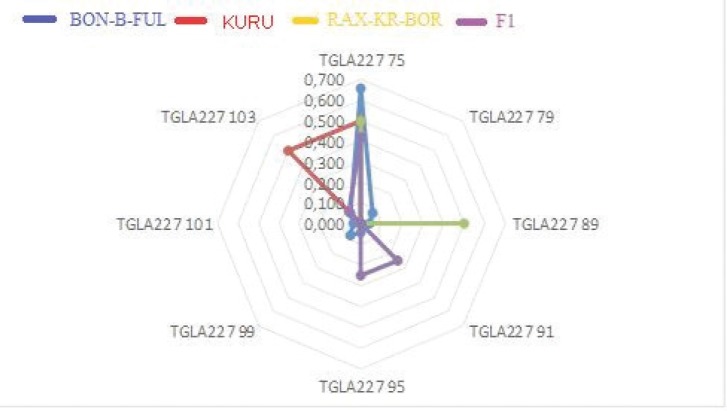
As shown in Figure 1, it is noted that in all studied rocks, the minimum number of alleles was found in the BM1824 locus—no more than three (F1). It is noteworthy that the number of effective alleles that make the main contribution to the calculation of the level of heterozygosity in the BON-B-FUL breed and in F1 crossbones was approximately the same, and is 3.98 ± 0.63 and 3.96 ± 0.45, respectively. The Shannon Information Index, which is a measure of species diversity is maximum for zebu of the White Fulani and F1 heifers born of them and is 1.44 ± 0.16 and 1.41 ± 0.11, respectively. It is worth noting that the number of private alleles exceeding the value of two was found only in the zebu of the White Fulani breed, which is probably due to the maximum sample size among all the studied populations (Fig. 1).
Figure 1. Graphical distribution of the frequencies of occurrence of MC alleles.
The study of allelic MS profiles allows us to trace the introduction of different breeds and Nigerian zebu generations involved in the formation of the F1 heifer genotype. Thus, the introduction of bulls of the Red Bororo, which probably served as a paternal basis in obtaining heifers F1, manifests itself in a change in the frequency of occurrence of the allele of the SPS115 locus: allele 246 from 50.0% to 25.0% and 248 from 50.0 up to 37.5% (in cows White Fulani, the frequency of occurrence of these alleles is 12.5% and 18.8%, respectively) and TGLA126 locus: allele 117 from 50.0% to 41.7% (in White Fulani cows, the frequency of occurrence of these alleles is 25.0%). As shown in the histograms for the SPS115 and TGLA126 loci, hybrid heifers F1, percentage of polymorphic loci d I BON-B-FULL and F1 was 100%, for KURU and RAX-KR-BOR—70% and 90%, respectively.
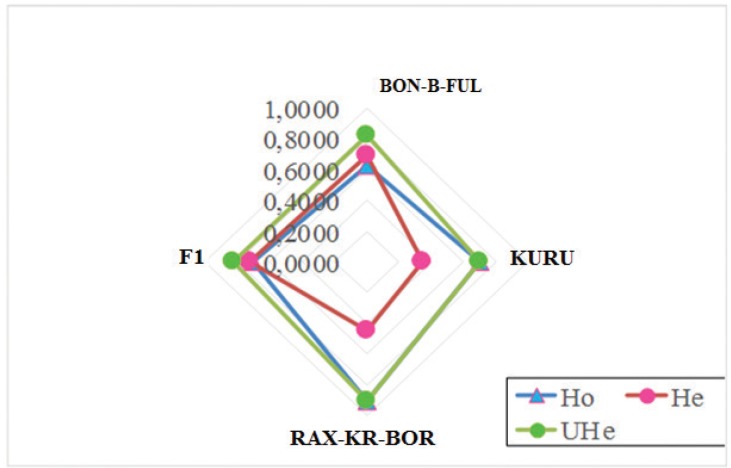
In order to estimate the variability in the zebu breeds studied, a calculation was made of the observed and expected degrees of heterozygosity. As is known, the heterozygosity index is a reflection of mutation processes, various types of selection, gene drift, non-random mating, and other factors of population dynamics [19]. The results of the analysis of heterozygosity levels are presented in Table 1.
As shown in Table 1 and Figure 2, the actual degree of heterozygosity ranged from 62.15% in BON-B-FUL zebu to 71.00% in F1 hybrids. A deficit of heterozygotes was found in these populations, with the highest value of the indicator observed in the BON-B-FUL breed —6.42%. The identified deficit of heterozygotes indicates the possible use of moderate inbreeding in breeding. The deficit of heterozygotes is also indicated by positive values of the Fis fixation index—from 0.019 in the F1 population (Fig. 2).
Figure 2. A graphic illustration of differences in heterozygosity levels is presented.
The results of studies on indicators of F-statistics are presented in Table 2. The Fis fixation index allows you to establish a connection between individuals of a particular breed and the entire sample as a whole. Since this indicator quantitatively reflects the deviation of the frequencies of heterozygous genotypes from the theoretically expected Hardy-Weinberg fraction of the proportion of heterozygotes during random mating within a population, it can be considered as one of the criteria for inbreeding population. At the same time, a positive value of the Fis index means a lack of heterozygotes in the given population, while a negative value of the index indicates an excess of heterozygotes.
Table 2. F-statistics of differences in heterozygosity.
| Locus | Fis | Fit | Fst |
|---|---|---|---|
| TGLA227 | −0.576 | −0.307 | 0.171 |
| BM2113 | −0.171 | 0.165 | 0.287 |
| ETH10 | −0.134 | 0.077 | 0.186 |
| SPS115 | −0.119 | 0.117 | 0.211 |
| TGLA126 | −0.515 | −0.222 | 0.193 |
| INRA23 | −0.240 | −0.012 | 0.184 |
| BM1818 | −0.231 | 0.049 | 0.227 |
| ETH225 | −0.756 | −0.321 | 0.248 |
| BM1824 | −0.333 | 0.495 | 0.621 |
| ETH3 | −0.235 | 0.012 | 0.200 |
| Total average for all breeds | −0.331 ± 0.068 | 0.005 ± 0.078 | 0.253 ± 0.042 |
The calculation of the Fis value showed an excess of heterozygotes in each individual microsatellite marker, and collectively in all animals. The Fit index establishes the relationship between individuals and the general array of animals studied. Similar to Fis, the Fit index indicates a deviation in the number of heterozygotes in the array under study from the theoretically expected Hardy-Weinberg. The calculation of the Fit value showed a 0.5% deficit of heterozygotes in the zebu breeds studied.
The Fst index serves as a criterion for genetic differences between breeds. This indicator always has a positive value, since the average degree of heterozygosity within a population, despite the randomness of pairing, is always lower compared to the entire array, since individuals within the population usually have common ancestors. The values of the Fst index lie between 0 (indicating the absence of genetic differences between populations) and 1 (indicates the fixation of specific alleles in the respective populations). The average Fst value for 10 loci for all breeds was 0.253. This suggests that 74.7% of all variability is due to intrabreed diversity and 25.3% to interbreeding diversity. The value of this indicator was higher compared with the cattle breeds studied earlier (Fst = 0.114), pigs (Fst = 0.152), and goats (Fst = 0.143), where the intra-breed diversity was 88.6%, 84.8%, and 85.7%, respectively. This indicates greater genetic isolation of zebu breeds from each other compared with cattle and pig breeds.
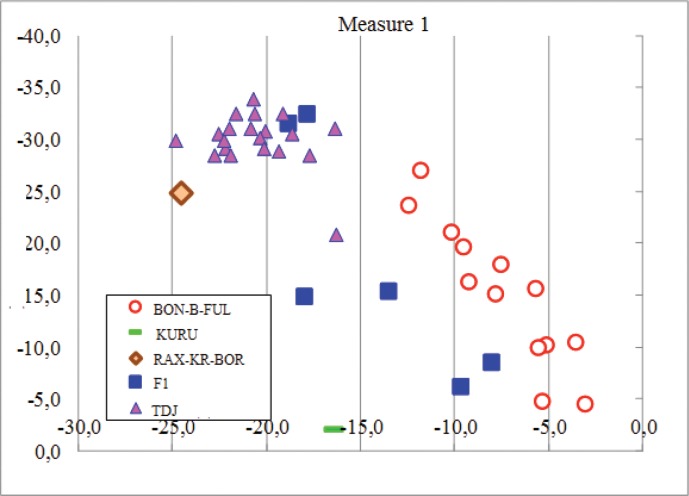
As shown in Figure 3, the determination of an individual’s genetic affiliation to its population, or in other words, the probability of identifying an individual’s pedigree affiliation based on its MS analysis, shows how far each population is from each other, subject to independent inheritance of microsatellites [19]. The analysis carried out on 10 MC loci showed a high identity of the individuals in the three breeds of 100% of the animal populations of KURU, PAX-KR-BOR, and F1.
Figure 3. Determining the belonging of individuals to a group.
As shown in Table 3 and in Figure 4, the greatest genetic distance from the studied zebu breeds was characterized by a breed of local zebu-like cattle in Tajikistan.
Table 3. Genetic distances between the studied zebu breeds, as calculated by Nei et al. [25].
| Breeds | BON-B-FUL | KURU | RAX-KR-BOR | F1 | TJ |
|---|---|---|---|---|---|
| BON-B-FUL | 0.000 | ||||
| KURU | 0.495 | 0.000 | |||
| RAX-KR-BOR | 0.459 | 0.425 | 0.000 | ||
| F1 | 0.531 | 0.560 | 0.554 | 0.000 | |
| TJ | 0.659 | 0.597 | 0.644 | 0.591 | 0.000 |
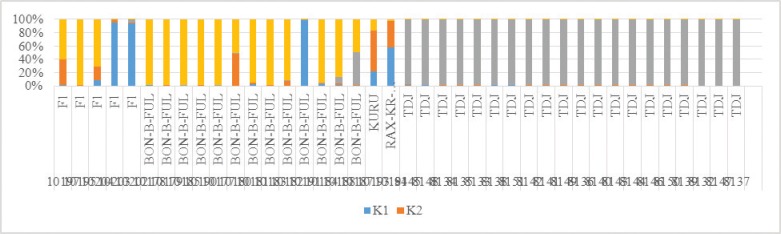
Figure 4. Differentiation of different cattle breeds, as described by Pritchard [26].
An assessment of the degree of differentiation performed by Pritchard [20] with k = 4 (the number of clusters corresponds to the number of the studied breeds) showed a clear differentiation of purebred Tajik zebuvid cattle (Q = 98.7%) and the main part of the White Fulani (Q = 81.8%). As shown in Figure 4, the results of the analysis of the data confirm the closest proximity of the populations and breed of Nigeria to each other, but also speak about the mixed origin of a number of individuals of the White Fulani breed, Kuru, and Red Bororo.
Thus, the results of our studies show that White Fulani is characterized by a high degree of genetic diversity. In the study of Ibeagha-Awemu and Erhardt [21], on genetic indices of the Red Bororo and white Fulani cattle breeds, they reported that the diversity was higher in the Cameroonian white Fulani. Present reports (Fig. 4) on a high degree of diversity in agreement with their findings on Cameroonian variety of cattle [21, 22].
In all studied breeds, the minimum number of alleles (not more than three in F1) was detected in the BM1824 locus. The number of effective alleles that make the main contribution to the calculation of the level of heterozygosity in the BON-B-FUL breed and in F1 hybrids was approximately equal and amounted to 3.98 ± 0.63 and 3.96 ± 0.45, respectively. The Shannon Information Index, which characterizes the measure of species diversity, is maximum for zebu of the white Fulani and heifers F1 (1.44 ± 0.16 and 1.41 ± 0.11, respectively).
It was established that the actual degree of heterozygosity in the studied populations varied from 62.15% in BON-B-FUL zebu to 71.00% in F1 hybrids. Simultaneously, a deficit of heterozygotes was found in the zebu populations, with the highest value of the indicator observed in the BON-B-FUL breed —6.42%. Present range is reported by Kasarapu et al. [23] in study on Bos taurus breeds. The presence of a deficit in heterozygotes suggests that moderate inbreeding was probably used when breeding the studied species. High identity of individuals of three breeds is shown: 100% of animal populations of KURU, PAX-KR-BOR, and F1. White Fulani breed cows for 92.3% of individuals correspond to their own population.
The greatest genetic distance according to Nei et al. [24], from the studied African zebu breeds, a breed of local zebu-like cattle of Tajikistan was characterized. White Fulani cows and their F1 descendants form a common cluster, to which the bulls of the Kuru and Red Bororo breeds are adjacent. The degree of differentiation by Pritchard [20] indicates a clear differentiation of purebred populations of Tajik zebu-like cattle (Q = 98.7%) and a significant proportion of White Fulani (Q = 81.8%) from Nigeria. At the same time, the cross origin of a number of white Fulani, Kuru, and red Bororo breeds is shown. Whereas, in our previous study, the phenotypic characterization of Tajk Zebu-like cattle was reported and described with differences from other similar breeds [7].
With attention to the finding of the present study and in according to Olutogun et al. [25], the White Fulani Zebu and their F1 crossbred heifers under similar farming and commercial environment is an efficient animal in lactation term. The sustainability of these cows for commercial milk production and also dual-propose farming under the tropical environment is documented in the present study, as described by Olutogun et al. [25]. In this study, it was not possible to take samples from all geographical areas. Studies of subsequent generations of this animal were also not possible due to time constraints. It is recommended for further studies.
Conclusion
Thus, the results of our studies show that White Fulani is characterized by a high degree of genetic diversity. White Fulani cows and their F1 descendants form a common cluster, to which the bulls of the Kuru and Red Bororo breeds are adjacent. There is a clear differentiation of purebred populations of Tajik zebu-like cattle (Q = 98.7%) and a significant proportion of white Fulani (Q = 81.8%) from Nigeria. At the same time, the cross origin of a number of white Fulani, Kuru, and red Bororo breeds is shown.
Acknowledgment
The authors are thankful to “The People’s Friendship University of Russia”, “All-Russian Research Institute for Animal Husbandry (VIZh),” and “Tajik Academy of Agricultural Sciences” for their help and support.
Conflict of interests
The authors declare that they have no conflict of interest.
Authors’ contribution
All of the authors have the same roles in the experimental process, analysis, writing, and editing the manuscript.
References
- [1].Glowatzki-Mullis M, Gaillard C, Wigger G, Fries R. Microsatellite-based parentage control in cattle. Anim Genet. 1995;26:7–12. doi: 10.1111/j.1365-2052.1995.tb02612.x. https://doi.org/10.1111/j.1365-2052.1995.tb02612.x. [DOI] [PubMed] [Google Scholar]
- [2].Lanari MR, Taddeo H, Domingo E, Centeno MP, Gallo L. Phenotypic differentiation of exterior traits in local Criollo goat population in Patagonia (Argentina) Arch Anim Breed. 2003;46:347–56. https://doi.org/10.5194/aab-46-347-2003. [Google Scholar]
- [3].Franklin IR. The utilization of genetic variation. Proc Assoc Advmt Anim Breed Genet. 1997;12:39–47. [Google Scholar]
- [4].Santoze A, Gicheha M. The status of cattle genetic resources in west africa: a review. Adv Anim Vet Sci. 2019;7(2):112–21. [Google Scholar]
- [5].Akpa GN, Alphonsus C, Abdulkareem A. Evaluation of herd structure of white Fulani cattle holdings in Zaria-Nigeria. Afr J Anim Biomed Sci. 2012;7(1):128–31. https://doi.org/10.5897/SRE11.458. [Google Scholar]
- [6].Mwai O, Hanotte O, Kwon YJ, Cho S. African indigenous cattle: unique genetic resources in a rapidly changing world. Asian-Austral J Anim Sci. 2015;28(7):911–21. doi: 10.5713/ajas.15.0002R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Norezzine A, Rebouh NY, Souadkia M, Parpura D, Gadzhikurbanov A, Gladyr EA, et al. Phenotypic characterization of the zebu cattle in Tajikistan. Int J Anim Vet Sci. 2018;12(1):22–6. [Google Scholar]
- [8].Ahmed K, Tamir B, Mengistu A. Cattle selection criteria and fattening practices in urban and peri-urban Kebeles of dessie and kombolcha towns, Ethiopia. Online J Anim Feed Res. 2017;7(2):29–37. [Google Scholar]
- [9].Kruglyak L. The use of a genetic map of biallelic markers in linkage studies. Nat Genet. 1997;17:21–4. doi: 10.1038/ng0997-21. https://doi.org/10.1038/ng0997-21. [DOI] [PubMed] [Google Scholar]
- [10].Landegren U, Nilsson M, Kwok PY. Reading bits of genetic information: Methods for single nucleotide polymorphism analysis. Genome Res. 1998;8:769–76. doi: 10.1101/gr.8.8.769. https://doi.org/10.1101/gr.8.8.769. [DOI] [PubMed] [Google Scholar]
- [11].Vignal A, Milan D, San Cristobal M, Eggen A. A review on SNP and other types of molecular markers and their use in animal genetics. Genet Sel Evol. 2002;34:275–305. doi: 10.1186/1297-9686-34-3-275. https://doi.org/10.1051/gse:2002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Heaton MP, Harhay GP, Benett GL, Stone RT, Grosse WM, Casas E, et al. Selection and use of SNP markers for animal identification and paternity analysis in U.S. beef cattle. Mammal Genome. 2002;13(5):272–81. doi: 10.1007/s00335-001-2146-3. https://doi.org/10.1007/s00335-001-2146-3. [DOI] [PubMed] [Google Scholar]
- [13].Werner FAO, Durstewitz G, Huberman FA, Thal ler G, Kramer W, Kollers S, et al. Detection and characterization of SNPs useful for identity control and parentage testing in major European dairy breeds. Anim Genet. 2004;35(1):44–9. doi: 10.1046/j.1365-2052.2003.01071.x. https://doi.org/10.1046/j.1365-2052.2003.01071.x. [DOI] [PubMed] [Google Scholar]
- [14].Williamson G, Payne WJA. 3rd. Longman Group; New York, NY: 1989. Introduction to Animal Husbandry in the Tropics. [Google Scholar]
- [15].Mbap ST, Bawa IA. Characterization of white Fulani and Sokoto Gudali cattle breeds in Bauchi State, Nigeria. Nigerian J Anim Prod. 1999;28(1):113–18. [Google Scholar]
- [16].Koger M. Characteristics of Types and Breeds of Cattle in the Tropics. In: Ristic M, McIntyre WIM, editors. Diseases of cattle in the tropics, current topics in veterinary medicine and animal science. Vol. 6. Springer; Dordrecht, The Netherlands: 1981. pp. 3–22. https://doi.org/10.1007/978-94-011-9034-3_1. [Google Scholar]
- [17].Nsoso BSJ, Aganga AA, Moganetsi BP, Tschwenyane SO. Birth weight, body condition score and heart girth in indigenous Tswana goats during dry and wet season in southeast Bostwana. Livest Res Rural Dev. 2003;15:32. [Google Scholar]
- [18].Brežná B, Piknová Ľ. Real-time PCR methods for identification of animal or plant species (chapter 18) In: David Rodríguez-Lázaro D, editor. Real-time PCR in food science: current technology and applications. Caister Academic Press; Poole, UK: 2013. pp. 255–74. [Google Scholar]
- [19].Boettcher P, oldenbroek K, Sponenberg D, Gandini G, Fernandez M, Joshi K. FAO Animal Production and Health Guidelines. No. 14. FAO; Rome, Italy: 2013. In vivo conservation of animal genetic resources; pp. 117–20. ISBN: 978-92-5-107725-2. [Google Scholar]
- [20].Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. https://doi.org/10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mariante A, Egit AA, Albuquerque M, Socorro M, Samuel R, Floriani RA. Managing genetic diversity and society needs. Rev Brasileira Zootecnia. 2008;37(spe):127–36. https://doi.org/10.1590/S1516-35982008001300016. [Google Scholar]
- [22].Ibeagha-Awemu EM, Erhardt G. An evaluation of genetic diversity indices of the Red Bororo and White Fulani cattle breeds with different molecular markers and their implications for current and future improvement options. Trop Anim Health Prod. 2006;38:431–41. doi: 10.1007/s11250-006-4347-y. https://doi.org/10.1007/s11250-006-4347-y. [DOI] [PubMed] [Google Scholar]
- [23].Kasarapu P, Porto-Neto LR, Fortes MRS, Lehnert SA, Mudadu MA, et al. The Bos taurus–Bos indicus balance in fertility and milk related genes. PLoS One. 2017;12(8):e0181930. doi: 10.1371/journal.pone.0181930. https://doi.org/10.1371/journal.pone.0181930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nei M, Tajima F, Tateno Y. Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. J Mol Evol. 1983;19(2):153–70. doi: 10.1007/BF02300753. https://doi.org/10.1007/BF02300753. [DOI] [PubMed] [Google Scholar]
- [25].Olutogun O, Yode-Owolade A, Abdullah AR. Comparative analysis of lactation traits of Holstein-Friesian, White Fulani Zebu and F1 crossbred cows in Nigeria; 8th World Congress on Genetics Applied to Livestock Production; August 13–18, 2006; Belo Horizonte, MG, Brazil. 2006. [Google Scholar]