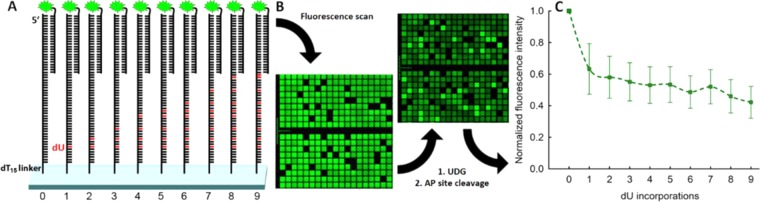
Figure 3.
(A) Sequence design for the investigation of uracil and abasic site cleavage on microarrays. Each sequence consists of a dT15-linker, then a 30mer with either no dUs (control) or an increasing number of dU-incorporations (from 1 to 9) replacing dTs in the following sequence: 5′-TTA CCA TAG AAT CAT GTG CCA TAC ATC ATC-3′. At the 5′-end, a control 25mer is synthesized (QC25), serving as target for the hybridization to its 3′-Cy3-labelled complementary oligonucleotide (QC25c). The cleavage process was monitored by recording the hybridization-based fluorescence intensity before and after the UDG-mediated cleavage of uracil nucleotides. (B) Small excerpt (ca. 7% of the total synthesis area) of fluorescence scans before and after enzyme exposure. The scans show the fluorescence intensity, resulting from hybridization to a labelled, complementary oligonucleotide. The microarrays were scanned at 5 µm resolution. (C) Decrease in fluorescence intensity for the UDG-mediated uracil excision (thus generating abasic sites) as a function of the number of dU nucleotide incorporations per DNA substrate. The actual cleavage efficiencies correlate with the loss of fluorescence intensity resulting from DNA substrate cleavage. The array was incubated for one hour with UDG and the generated abasic sites were subsequently cleaved under alkaline conditions. The decrease in fluorescence intensity was recorded and normalized to the control strand (U0). The normalized intensities, indicated in arbitrary units, were plotted over the number of dUs per DNA substrate. Error bars are SD.