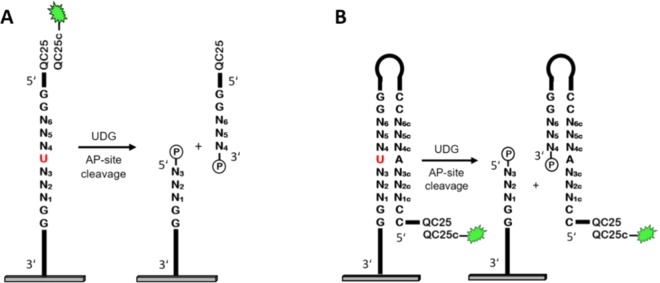
Figure 5.
Schematic illustration of the sequence design for the investigation of E. coli UDG sequence dependences on single- (A) and double-stranded DNA substrates. (B) In order to investigate the UDG sequence dependence, a single dU is incorporated into a DNA strand and enclosed by 3 permuted bases on each side. A The design for the study of UDG sequence dependence on single-stranded DNA substrates consists of a 15mer dT-linker, a single dU enclosed by 3 permuted bases on each side, followed by a 5′ 25mer sequence (QC25) serving as target for the hybridization to its 3′-Cy3-labelled complementary oligonucleotide (QC25c). B For the study of UDG sequence dependence on double-stranded DNA substrates, the sequences were designed to form a hairpin loop. The resulting strands consisted of a 15mer dT-linker, a 11-nt stem, equivalent to the single-stranded design, containing the variable region flanked by dG·dC base pairs, a 4-nt loop followed by the complementary 11nt strand. At the 5′ end, a hybridizable 25mer target sequence (QC25) was synthesized.