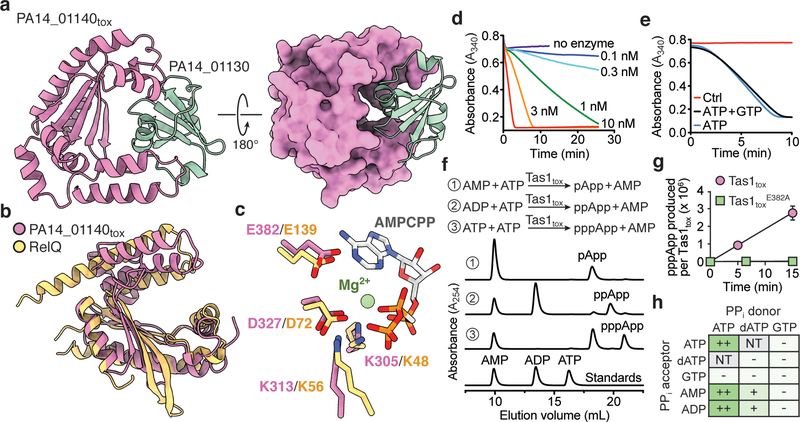
Figure 2 |. Tas1tox adopts a RelA-SpoT Homolog (RSH) fold found in enzymes that synthesize the bacterial alarmone (p)ppGpp but instead synthesizes (p)ppApp.
a) Overall structure of PA14_01140tox in complex with PA14_01130. Shown are ribbon (left) and space-filling (right) representations of PA14_01140tox (purple) in complex with PA14_01130 (green). b) PA14_01140tox resembles (p)ppGpp synthetase enzymes. Structural overlay of PA14_01140tox and the small alarmone synthetase RelQ from Bacillus subtilis (PDB code 5DEC)22. The structures superimpose with a Cα r.s.m.d. of 3.4Å over 145 equivalent positions. c) Structural alignment of the pyrophosphate donor ATP binding site of RelQ in complex with a magnesium ion and the non-hydrolyzable ATP analog AMPCPP (PDB code 5F2V) with the equivalent amino acid positions in PA14_01140tox. Amino acid side chains deriving from PA14_01140tox or RelQ and their corresponding labels are shown in purple and yellow, respectively. d) PA14_01140tox catalyzes the formation of AMP in a dose-dependent manner. Coupled enzyme assay of PA14_01140tox-catalyzed AMP production as a function of NADH consumption over time. e) PA14_01140tox catalyzes the production of AMP from ATP in a GTP-independent manner. The control reaction lacks adenylate kinase, which is required for the initial step of the coupled assay. f) PA14_01140tox (Tas1tox) is a (p)ppApp synthetase enzyme. Anion-exchange traces of ATP alone or with excess AMP or ADP after incubation with Tas1tox. A standard trace for ATP, ADP and AMP is shown for comparison. g) Rate of ppApp production by Tas1tox or Tas1toxE382A. Reactions were performed at 37°C with 10 mM ATP and 1 nM Tas1tox or 1 μM Tas1toxE382A. Data are mean ±SD from three separate reactions. h) Specificity of Tas1tox towards pyrophosphate (PPi) donors and acceptors. Indicated nucleotides (1 mM each) were incubated with 100 nM Tas1tox at room temperature for 10 minutes. Reactions that progressed to completion (++), made detectable product (+) or made no detectable product (−) are indicated. NT, not tested. See also Extended Data Figure 6. d–f) Data are representative of two technical replicates.