Figure 4 |. (p)ppApp interacts with PurF and inhibits de novo purine biosynthesis.
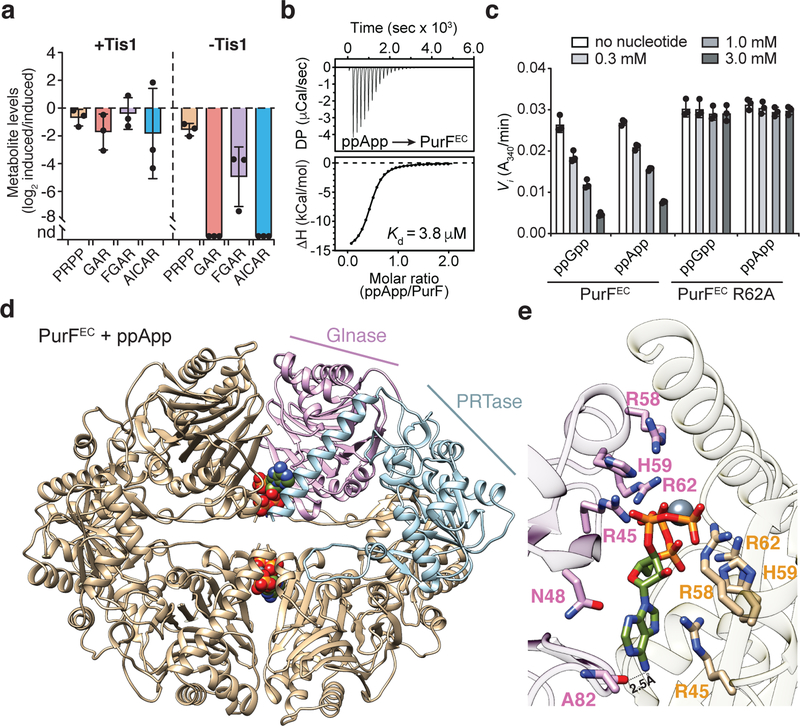
a) Relative quantification of metabolites within the de novo purine biosynthesis pathway in P. aeruginosa strains containing or lacking Tis1. Metabolite levels for both the +Tis1 and –Tis1 strains are shown as log2 ratios for samples 1-hour post-induction relative to pre-induction of sspB expression. nd, not detected. b) Isothermal calorimetry traces (top) and fitted isotherms (bottom) for the titration of 100 μM PurFEC (monomer) with 1 mM ppApp. Representative traces from two independent replicates are shown. c) Changes to the activity of PurFEC in the presence of indicated concentrations of ppGpp or ppApp. d) Ribbon diagram of the PurFEC tetramer in complex with ppApp. A single PurF subunit is coloured by individual domains (Glnase; glutaminase domain in pink, PRTase; phosphoribosyltransferase domain in blue), while the remaining subunits are coloured brown. ppApp and Mg2+ are shown in stick and sphere representations, respectively. e) Close-up view of the ppApp binding site between the glutaminase domains of two adjacent PurFEC monomers. ppApp-interacting residues are shown as pink or orange sticks and hydrogen bonding between PurFEC and the purine ring of ppApp are shown as black dashed lines (see Extended Data Fig. 10 for additional details). a, c) Data are mean ±SD for three separate cultures (a) or reactions (c).
