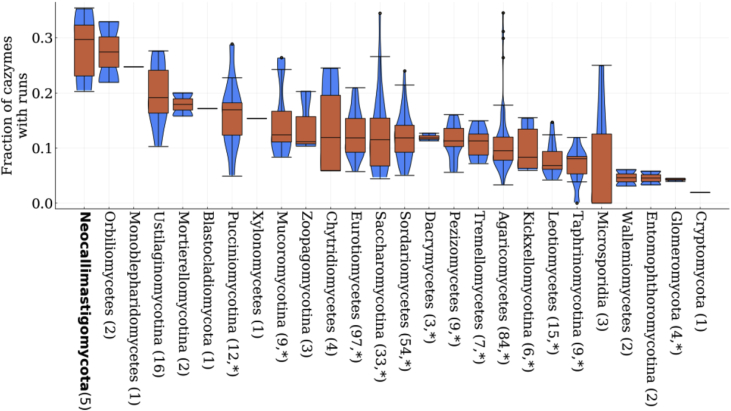
Fig. 4.
Neocallimastigomycota have significantly more CAZymes with amino acid repeatrunsthan other fungal clades. Here the distribution of the fraction of CAZymes with runs relative to all the CAZymes in each fungus, grouped by clade, is plotted. The number in parentheses is the number of fungi included in each clade. Statistically significant differences in the distributions between Neocallimastigomycota and all the other clades are indicated by * using the two sample Kolmogorov-Smirnoff test (P < 0.05). The distribution of the fraction of CAZymes with runs in each clade is shown in the blue violin plots overlaid by orange box-and-whisker plots where outliers are shown as points, minima and maxima as whiskers, and the inter-quartile ranges inside the boxes. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)