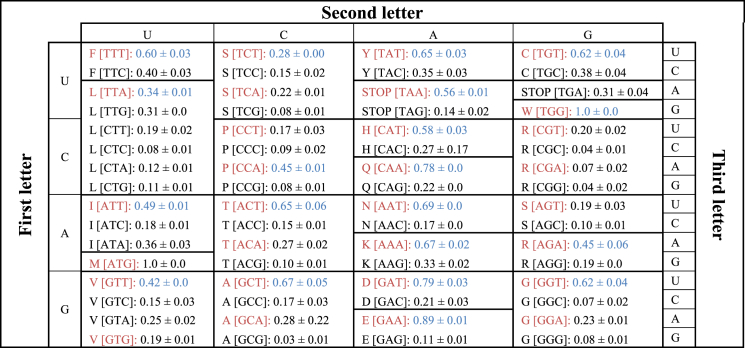
Table 2.
Codon optimization table for Neocallimastigomycota. Fraction of the proteome encoded for by each codon in highly expressed transcripts of N. californiae and A. robustus averaged, with standard deviation noted. The most AT rich codon for each amino acid is shown in red font, while the most abundant codon within the transcriptome is shown in blue font. AT-rich codons are invariably preferred in anaerobic fungi, in line with the predicted low GC content of the clade.