Fig. 3.
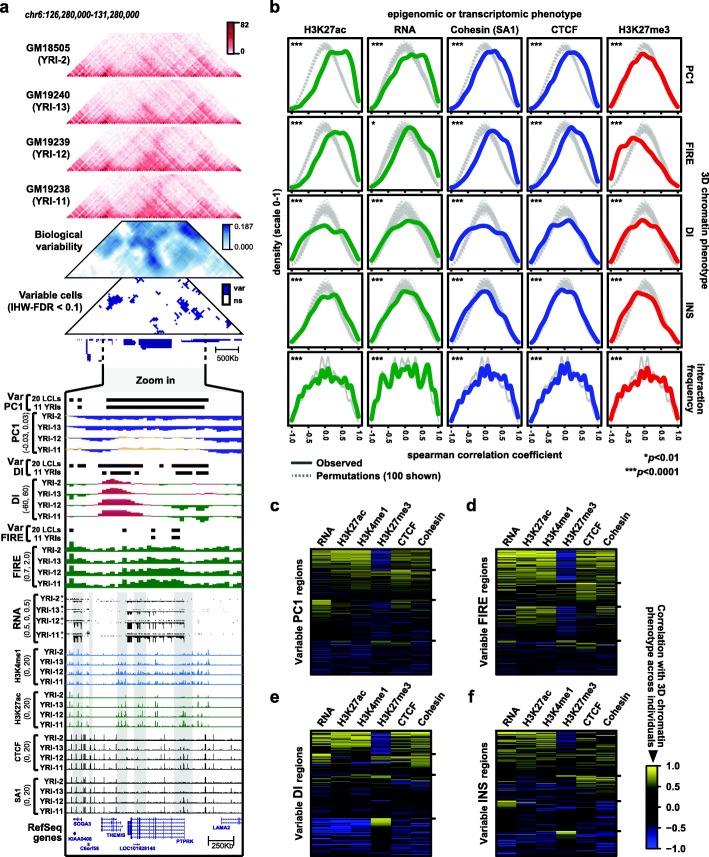
Coordinated variation of the 3D genome, epigenome, and transcriptome. a Example of a variable region where 3D chromatin phenotypes are correlated with epigenomic and transcriptomic phenotypes (chr6:126,280,000-131,280,000; hg19). Six triangular heatmaps from top to bottom: Hi-C contact heatmaps from four individuals, variability matrix, and variable cells in the matrix (var = variable, ns = not significant). Standard tracks below show 3D chromatin, epigenomic, and transcriptomic properties from four individuals in zoomed region (chr6:127,680,918-129,416,097; hg19). All ChIP-seq and RNA-seq data in the Figure from Kasowski et al. [42]. b Density plots show the distribution of Spearman correlation coefficients at variable regions between the epigenomic or transcriptomic phenotype indicated in the top margin and the 3D chromatin phenotype indicated in the right margin of panel. Gray lines show the distributions from 100 random permutations selected at random from the 10,000 permutations performed (due to plotting limitations). ***p < 0.0001 by permutation test as described in the “Identifying biological variability in Hi-C contact matrices” section, which applies to all observations in this panel except RNA-seq at INS regions (p = 0.0018) and RNA-seq at FIRE regions (p = 0.0096). c Heatmap showing Spearman correlation coefficients between PC1 and multiple epigenomic/transcriptomic phenotypes, arranged by k-means clustering (k = 4). Tick marks to the right show boundaries between clusters. Each row (N = 518) is one variable PC1 region, limited to the subset of variable PC1 regions that contain RNA-seq signal and at least one peak in at least one individual for each ChIP-seq target included here (H3K27ac, H3K4me1, H3K27me3, CTCF, Cohesin). d Similar to c, showing correlations with FIRE at N = 132 variable FIRE regions. e Similar to c, showing correlations with DI N = 265 variable DI regions. f Similar to c, showing correlations with INS at N = 154 variable INS regions