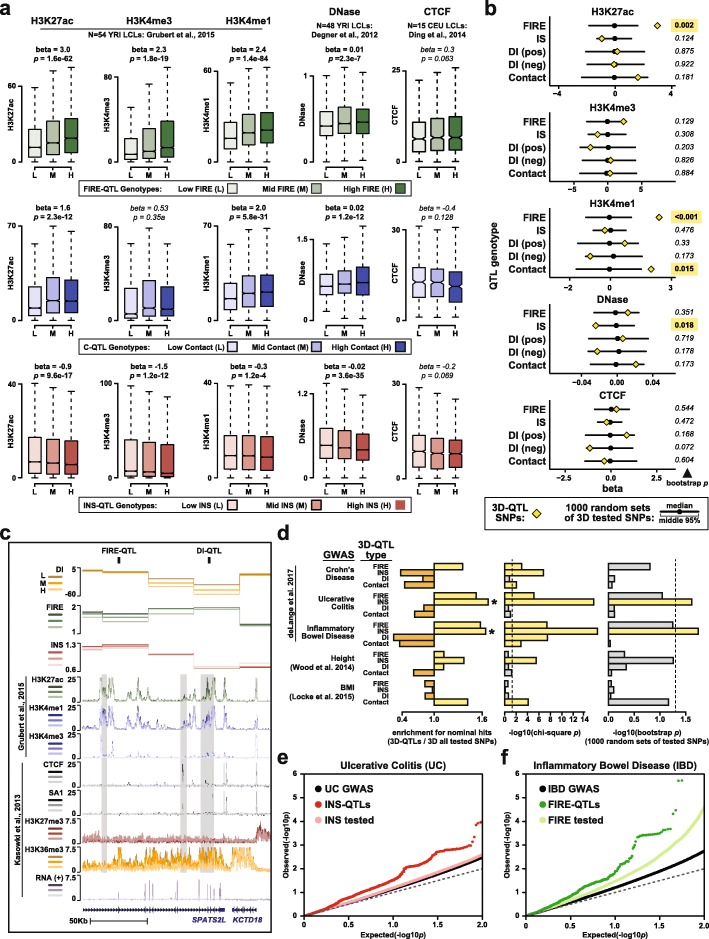
Fig. 5.
Contribution of 3D chromatin QTLs to other molecular and organismal phenotypes. a Boxplots show signal for epigenetic phenotypes separated by genotype at FIRE-QTLs (top row), C-QTLs (middle row), and INS-QTLs (bottom row). Epigenetic signals averaged across all peaks in 40-Kb bin. Linear regression p and beta values shown above each plot. p value < 0.05 in bold, others in italics. b Line plots shows beta values of linear relationships between QTL genotypes indicated to the left and epigenetic phenotype indicated above each subpanel. Yellow diamonds show beta values for the true QTLs sets as shown in (a) or Additional file 1: Figure S10b. Black circles and lines indicate the median and middle 95% range, respectively, of 1000 permutations in which SNPs were selected from those tested in the QTL search indicated to the left of each line. The number to the right of each line indicates the bootstrap p value (fraction of permutations with abs(beta) higher than observed for the true QTL set). Yellow boxes highlight values < 0.05. c Genome browser view (chr2:201,222,342-201,386,844; hg19) showing examples of a DI-QTL (chr2:201333312) and FIRE-QTL (chr2:201254049). All signals plotted as a function of DI-QTL genotype (L = Low DI, M = medium DI, H=High DI). Gray boxes highlight regions where epigenetic signals stratify by DI-QTL genotype. d Left subpanel shows the enrichment values for 3D QTL SNPs with nominal significance in the indicated GWAS study calculated as follows: (fraction of indicated 3D QTL SNPs with nominal significance in the indicated GWAS)/(fraction of 3D test SNPs with nominal significance in the indicated GWAS). Asterisks mark values with p < 0.05 by chi-square test (middle panel), and permutation test (right panel). Right panel shows the proportion of 1000 random SNP subsets (selected from the tested SNPs) with enrichment values higher than the indicated true QTL set. Dotted lines mark p < 0.05. e QQ plot shows the results of UC GWAS with all tested SNPs shows as black points, and two subsets as follows: SNPs also tested in our INS-QTL search (light red), and SNP called as INS-QTLs or in perfect LD with INS-QTLs in the same 40-Kb bin (dark red). f QQ plot shows the results of IBD GWAS with all tested SNPs shows as black points, and two subsets as follows: SNPs also tested in our FIRE-QTL search (light green), and SNP called as FIRE-QTLs or in perfect LD with FIRE-QTLs in the same 40-Kb bin (dark green)