Significance
Atrial fibrillation (AF) is the most common cardiac arrhythmia, affecting 2 to 3% of the general population and leading to significant morbidity and mortality. Genome-wide association studies identified the 4q25 AF risk locus, a region that forms long-range interactions with the promoter of PITX2, which encodes a critical transcriptional regulator of cardiac development. Using a zebrafish pitx2c loss-of-function model, we find that larval and adult zebrafish phenocopy many hallmarks of human AF. Our data further indicate that the pathogenesis of arrhythmia and AF-like phenotypes in pitx2c mutants is driven by developmental perturbations to sarcomere organization and metabolic pathways. We also find that antioxidant treatment reduces the incidence and severity of cardiac arrhythmia, suggesting avenues for therapeutic strategies.
Keywords: cardiac development, cardiomyopathy, cardiac metabolism, transcriptional profiling
Abstract
Atrial fibrillation (AF) is the most common type of cardiac arrhythmia. The major AF susceptibility locus 4q25 establishes long-range interactions with the promoter of PITX2, a transcription factor gene with critical functions during cardiac development. While many AF-linked loci have been identified in genome-wide association studies, mechanistic understanding into how genetic variants, including those at the 4q25 locus, increase vulnerability to AF is mostly lacking. Here, we show that loss of pitx2c in zebrafish leads to adult cardiac phenotypes with substantial similarities to pathologies observed in AF patients, including arrhythmia, atrial conduction defects, sarcomere disassembly, and altered cardiac metabolism. These phenotypes are also observed in a subset of pitx2c+/− fish, mimicking the situation in humans. Most notably, the onset of these phenotypes occurs at an early developmental stage. Detailed analyses of pitx2c loss- and gain-of-function embryonic hearts reveal changes in sarcomeric and metabolic gene expression and function that precede the onset of cardiac arrhythmia first observed at larval stages. We further find that antioxidant treatment of pitx2c−/− larvae significantly reduces the incidence and severity of cardiac arrhythmia, suggesting that metabolic dysfunction is an important driver of conduction defects. We propose that these early sarcomere and metabolic defects alter cardiac function and contribute to the electrical instability and structural remodeling observed in adult fish. Overall, these data provide insight into the mechanisms underlying the development and pathophysiology of some cardiac arrhythmias and importantly, increase our understanding of how developmental perturbations can predispose to functional defects in the adult heart.
The most prevalent defect affecting cardiac conduction is atrial fibrillation (AF), a common sustained arrhythmia associated with increased risk of cardioembolic stroke or sudden death. Despite its clinical importance, mechanisms underlying the initiation and pathogenesis of AF remain poorly understood. The genetic component of AF has been investigated through large-scale genome-wide association studies. The most prominently associated region is 4q25, an intergenic region with at least four independent AF association loci in close proximity to the PITX2 gene (1–4). A role for PITX2 in AF pathogenesis has been demonstrated by animal model studies (5–10). More recently, chromosome conformation capture (3C) studies have shown that 4q25 interacts with the promoter of the cardiac-specific isoform of PITX2C, further supporting the hypothesis that 4q25 variants regulate PITX2C expression (11).
PITX2 is a transcription factor that plays critical roles during cardiac morphogenesis. Three isoforms are generated from two promoters and alternative splicing. PITX2C is the major isoform expressed in the heart, with enrichment in the left atrium. It has previously been observed that PITX2 messenger RNA (mRNA) levels are reduced in the atrial cardiomyocytes (CMs) of sustained AF patients carrying risk variants at 4q25 (12). Additionally, PITX2 gain-of-function variants have also been associated with AF in humans, suggesting that expression levels of PITX2 targets must be tightly regulated for cardiac homeostasis (13, 14). During vertebrate development, Pitx2 is a key regulator of left–right patterning of the body plan (15–17). Within the conduction system, Pitx2 seems to play a similar role to repress left-sided pacemaker identity (5, 7, 9). Adult Pitx2+/− mice are prone to AF when paced, likely as a developmental patterning defect arising from expanded pacemaker gene expression (5, 7). Conditional genetic mouse models reveal a postnatal function for Pitx2 in directly regulating genes encoding ion channels and cell–cell junctions, thereby affecting cardiac action potentials (18).
Mechanisms of AF initiation and pathogenesis are not well understood. Developmental defects have been proposed to predispose to AF as indicated by association with developmental genes, such as PITX2. These defects are thought to result in subclinical abnormalities in the adult heart that can subsequently manifest as AF after induction by aging, disease, or stress. Additionally, mutations in genes required to maintain CM structure, such as titin-truncating variants and lesions in the atrial-specific myosin light chain gene MYL4, have been shown to predispose to AF (19–21), indicating that the disease is not only caused by electrical disturbances.
Here, we show that pitx2c−/− adult zebrafish display many pathologies found in human AF patients, including atrial conduction defects, general structural remodeling, and altered cardiac metabolism. Using transcriptomics approaches, we find that Pitx2c is a critical regulator of sarcomeric and metabolic gene expression during embryogenesis. Notably, we show that treatment with the antioxidant N-acetyl cysteine (NAC) can rescue cardiac arrhythmia in pitx2c−/− larvae, suggesting that altered mitochondrial-derived oxidative stress is an important driver of arrhythmia in conjunction with abnormal sarcomere organization. These data indicate that developmental perturbations to sarcomere function and metabolic pathways contribute to arrhythmia and AF-like pathologies.
Results
Loss of Pitx2c Leads to Altered Cardiac Function and Cardiomyopathy in Adult Zebrafish.
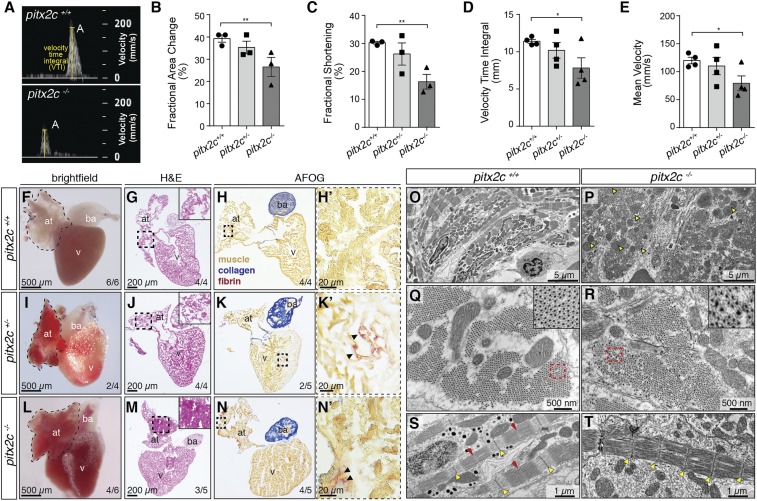
We investigated a recently generated loss-of-function allele of pitx2c (pitx2cups6) in zebrafish (22) and assessed its adult cardiac phenotype. pitx2cups6/up6 (hereafter referred to as pitx2c−/−) are fertile and survive until adulthood; however, increased mortality rates are observed compared with pitx2c+/+ siblings (SI Appendix, Fig. S1A). To evaluate whether cardiac function was affected, we first performed echocardiography on adult fish (Fig. 1 A–E). We quantified parameters of cardiac function by Brightfield-Mode (B-Mode) imaging as well as pulsed-wave Doppler imaging of blood flow. B-Mode imaging of sagittal long-axis views revealed cardiac dysfunction in pitx2c−/− compared with pitx2c+/+ siblings (Movies S1 and S2). From these views, we measured ventricular area and length during systole and diastole to quantify fractional area change (FAC) and fractional shortening (FS), respectively. pitx2c−/− adults exhibit significantly reduced FAC and FS (26.5 ± 0.043%; 16.3 ± 0.026%, respectively) compared with pitx2c+/+ siblings (39.3 ± 0.016%, P = 0.0495; 30.3 ± 0.005%, P = 0.0065). Quantification of peak ejection blood flow parameters (Fig. 1A) revealed a reduction in ventricular velocity time integral (VTI) (Fig. 1D) and mean ventricular velocity (Fig. 1E) in pitx2c−/− (11.4 ± 0.26 mm; 120.0 ± 5.8 mm/s, respectively) compared with pitx2c+/+ siblings (7.8 ± 1.4 mm, P = 0.0428; 79.17 ± 13.2 mm/s, P = 0.0296). Together, these data indicate impaired cardiac function in pitx2c−/− adult zebrafish.
Fig. 1.
Loss of Pitx2c disrupts cardiac function and structure in adult zebrafish. (A–E) Analysis of cardiac function by echocardiography. Brightfield-Mode (B-Mode) imaging of long-axis views was used to quantify FAC and FS. Pulsed-wave Doppler imaging (A) was used to quantify peak A VTI and mean blood flow velocity (E). Individual data points represent an average measurement taken from 3 diastolic/systolic phases of each fish. All parameters of cardiac function are significantly reduced in pitx2c−/− adults, with variability observed in pitx2c+/− siblings. (F–N) Adult cardiac morphology at 14 months postfertilization (mpf). Dashed lines outline the atrium in whole-mount view of hearts (F, I, and L). Histology was analyzed by hematoxylin and eosin (H&E) staining (G, J, and M; Insets show higher magnification of the boxed region), and fibrosis was analyzed by Acid Fuchsin Orange G (AFOG) staining (H, K, and N; higher magnification of the boxed regions are shown in H’, K’, and N’). pitx2c−/− hearts display enlarged and blood-filled atria (L), and atrial tissue appears hypertrophic (M, Inset). Fibrotic foci are present in the atrium of pitx2c−/− fish (N′; black arrowheads). Intermediate phenotypes were observed in pitx2c+/− hearts. at, atrium; ba, bulbus arteriosus; v, ventricle. (O–T) TEM images of adult pitx2c+/+ and pitx2c−/− atria (n = 3). Areas of highly damaged and necrotic tissue are observed in pitx2c−/− atria, with an increased number of mitochondria that appear enlarged and defective (P; yellow arrowheads). Representative images of cross-sections through myofibril bundles (Q and R; Q, Inset and R, Inset show higher magnifications) and longitudinal sections of sarcomeres (S and T). pitx2c−/− atria exhibit disorganized myofibril bundles and poorly defined Z discs (yellow arrowheads), and M lines (red arrows) are difficult to distinguish (T). Error bars indicate SEM. *P < 0.05 by unpaired t test; **P < 0.01 by unpaired t test.
We next analyzed gross cardiac morphology. On dissection of adult hearts, we found that a subset of pitx2c−/− atria was enlarged and remained filled with blood following perfusion with saline (Fig. 1L). Subsequent histological analysis of pitx2c−/− atria revealed hypertrophic regions (Fig. 1M) as well as focal areas of collagen and fibrin staining, suggesting increased fibrosis (Fig. 1 N and N′). To confirm the presence of fibrosis, we measured mRNA levels of the profibrotic genes acta2 and col1a1a by qPCR and observed their up-regulation in pitx2c−/− hearts compared with pitx2c+/+ (SI Appendix, Fig. S1B). We noted similar phenotypes in a subset of pitx2c+/− adults (∼50%) (Fig. 1 I–K). We did not detect obvious fibrosis in young adult hearts (SI Appendix, Fig. S1 C–H), suggesting that fibrosis likely arises as a secondary consequence of altered cardiac function.
To examine cardiac morphology at the ultrastructural level, we performed transmission electron microscopy (TEM) on atria of pitx2c+/+ and pitx2c−/− adult fish (Fig. 1 O–T). Atrial tissue architecture in pitx2c−/− was heterogenous, with extensive areas of necrosis and tissue damage (Fig. 1P). Mitochondrial morphology was also variable, with many smaller dilated mitochondria observed in pitx2c−/− atria (Fig. 1P). Cross-sections through sarcomeres in pitx2c+/+ atria revealed parallel arrangement of thick and thin filaments forming a tight hexagonal array (Fig. 1Q), with clearly defined Z discs and M lines that anchor myofibrils in place (Fig. 1S). In contrast, myofibrils were disorganized and irregularly spaced in pitx2c−/− atria, and Z discs and M lines were poorly defined (Fig. 1 R and T). These data suggest that loss of Pitx2c leads to cardiomyopathy phenotypes in adult zebrafish.
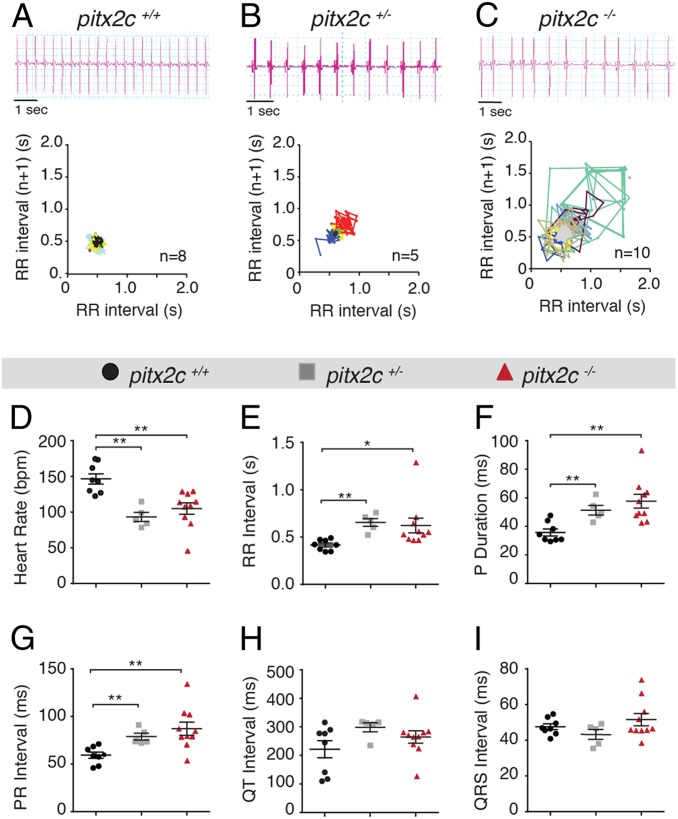
Next, we performed electrocardiogram (ECG) analysis (Fig. 2) to assess cardiac electrophysiology. Cardiac arrhythmia was observed in ECG traces from pitx2c+/− and pitx2c−/− animals (Fig. 2 B and C) as well as prolonged RR intervals (the time between ventricular depolarizations) (Fig. 2E). We also observed a reduction in the average heart rate in both pitx2c+/− (93.3 ± 6.3 bpm) and pitx2c−/− (105.0 ± 7.9 bpm) fish compared to pitx2c+/+ siblings (146.4 ± 7.2 bpm) (Fig. 2D). P-wave duration (Fig. 2F) and PR intervals (representing atrial depolarization) (Fig. 2G) were significantly increased in pitx2c+/− (51.33 ± 3.34 ms; 78.91 ± 3.62 ms, respectively) and pitx2c−/− (57.61 ± 4.83 ms; 87.13 ± 6.93 ms) fish compared with pitx2c+/+ siblings (35.75 ± 2.43 ms; 59.30 ± 3.20 ms, respectively), indicating atrial conduction defects. In contrast, QT and QRS intervals, the time from ventricular contraction until relaxation and ventricular depolarization, respectively, were not significantly different between the genotypes (Fig. 2 H and I). Together, these data indicate that loss of Pitx2c leads to atrial conduction defects without affecting the ventricular conduction system.
Fig. 2.
Cardiac arrhythmia and prolonged atrial depolarization in pitx2c−/− adult hearts. Electrical conduction properties of 8-month postfertilization (mpf) adult hearts by ECG analysis. Representative ECG traces and Poincaré plots of consecutive RR intervals from pitx2c+/+ (A), pitx2c+/− (B), and pitx2c−/− (C) hearts. Individual fish are each represented by a differently colored line. Quantification of heart rate (D), RR interval (E), P-wave duration (F), PR interval (G), QT interval (H), and QRS interval (I). pitx2c+/− and pitx2c−/− fish exhibit a variable and lower heart rate (B–E) and disrupted atrial conduction (F and G), while the ventricular conduction system seems unaffected (H and I). Error bars correspond to SEM. *P < 0.05 by 1-way ANOVA; **P < 0.01 by 1-way ANOVA.
Loss of Pitx2c Function Leads to Mitochondrial Defects in the Adult Heart.
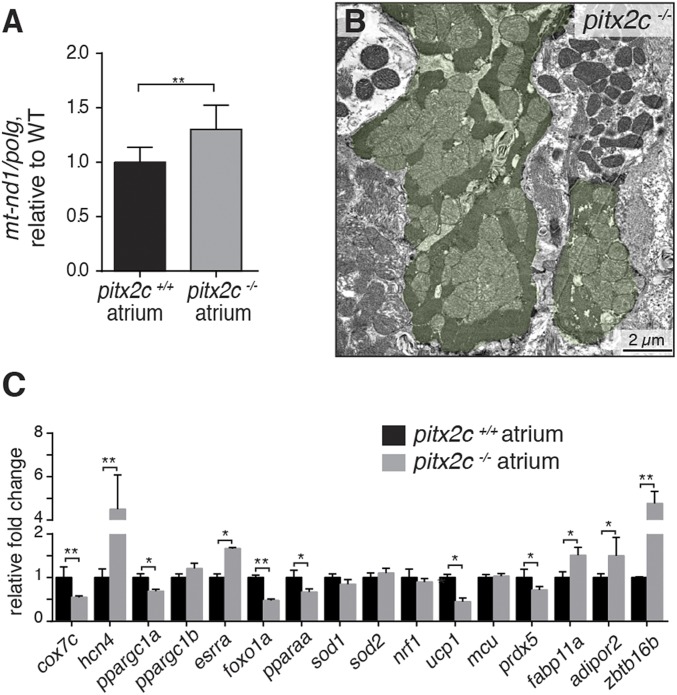
Cellular metabolic conditions are closely linked to the high-energy demand of the cardiac contractile machinery. Accordingly, altered cardiac energetics have been reported in human and experimental AF conditions (23, 24). The mitochondrial phenotypes observed by TEM (Fig. 1P) led us to hypothesize that cardiac metabolism was compromised in pitx2c−/− hearts. To test this hypothesis, we measured mitochondrial respiration and reactive oxygen species (ROS) production. While overall respiration was slightly increased in pitx2c−/− atria (SI Appendix, Fig. S2), ROS/O2 levels were lower in pitx2c−/− atria (0.004 ± 0.001 pmol H2O2/JO2) compared with pitx2c+/+ siblings (0.014 ± 0.012 pmol H2O2/JO2) (SI Appendix, Fig. S2). In contrast, we observed reduced maximal mitochondrial respiration (Electron Transport System State 3) in pitx2c−/− ventricles (76.7 ± 14.5 pmol O2) compared with pitx2c+/+ siblings (98.7 ± 16.7 pmol O2) (SI Appendix, Fig. S2), suggesting reduced oxidative phosphorylation capacity. To determine whether the alterations in respiration could result from changes in mitochondrial mass, we measured mitochondrial DNA (mtDNA) content in adult atria. mtDNA content was increased by ∼30% in pitx2c−/− atria compared with pitx2c+/+ siblings (Fig. 3A). These data are in line with the abnormal mitochondrial morphology observed in pitx2c−/− hearts (Fig. 1P), including the large mitochondrial clusters (Fig. 3B).
Fig. 3.
Mitochondrial and metabolic changes in pitx2c−/− adult atria. (A) Quantification of relative mtDNA content in pitx2c+/+ and pitx2c−/− atria by qPCR. (B) Representative image of mitochondrial clusters (highlighted in green) observed in pitx2c−/− adult hearts by TEM (n = 3). (C) qPCR of known PITX2 targets as well as additional metabolic genes in pitx2c+/+ and pitx2c−/− adult atria. Significant down-regulation of cox7c, ppargc1a, foxo1a, pparaa, ucp1, and prdx5 mRNA levels and up-regulation of hcn4 and esrra as well as adipogenic genes fabp11a, adipor2, and zbtb16b mRNA levels are observed in pitx2c−/− atria (C). Error bars correspond to SEM. *P < 0.05 by unpaired t test; **P < 0.01 by unpaired t test.
Direct Pitx2 target genes identified in neonatal mouse cardiac regeneration models include regulators of mitochondrial function, oxidation–reduction, and the respiratory chain (18, 25). To test whether these genes are regulated by Pitx2c in adult zebrafish hearts, we examined mRNA levels in atrial tissue by qPCR (Fig. 3C). We first examined the expression of the previously identified direct targets hcn4 and cox7c (9, 18). As reported in mouse, we observed up-regulation of hcn4 and down-regulation of cox7c in pitx2c−/− atria compared with pitx2c+/+ siblings. We then screened a panel of genes involved in cellular metabolism. pitx2c−/− atria exhibited significant reduction in ppargc1a, foxo1a, pparaa, ucp1, and prdx5 mRNA levels and up-regulation of esrra, fabp11a, adipor2, and zbtb16b mRNA levels (Fig. 3C). Together, these gene expression changes indicate alterations in oxidative phosphorylation and redox status of the heart as well as increased adipogenesis.
Pitx2c Controls Cardiac Rhythmicity in Zebrafish Larvae.
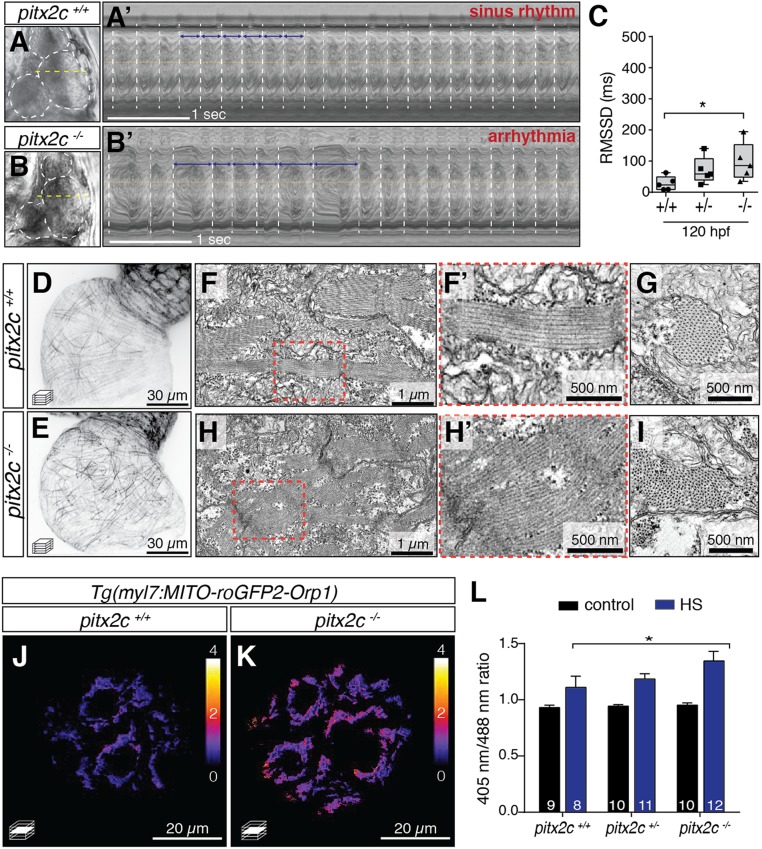
One of the major questions surrounding AF initiation is whether changes to developmental genes can predispose to AF manifestation later in life. Therefore, we analyzed the phenotypic onset in pitx2c−/− animals by testing for cardiac arrhythmia during developmental stages. To this end, we performed high-speed spinning disc confocal imaging to record bright-field movies of beating larval hearts (Movies S3 and S4) and plotted kymographs to visualize and quantify cardiac intervals (Fig. 4 A and B). Larvae were scored as sinus rhythm (Fig. 4 A and A′) or arrhythmia (Fig. 4 B and B′) based on the absence or presence of irregular cardiac intervals, respectively. We found that a subset of pitx2c−/− larvae was arrhythmic starting at 120 h postfertilization (hpf), and phenotype penetrance increased with age (SI Appendix, Fig. S3). We then quantified heart rate variation in zebrafish larvae at 120 hpf by calculating the root mean square of the successive differences (RMSSD) of cardiac cycles, a method used to assess short-term heart rate variability in humans (26). As RMSSD measurements reflect beat-to-beat variability, elevated RMSSD values indicate arrhythmic events. Indeed, quantification of RMSSD indices revealed elevated beat-to-beat variability in pitx2c−/− larvae (95.77 ± 60.62 ms) compared with pitx2c+/+ siblings (35.09 ± 20.90 ms) (Fig. 4C). Therefore, we conclude that loss of pitx2c leads to cardiac arrhythmia during early larval stages.
Fig. 4.
Cardiac arrhythmia in pitx2c−/− larvae is preceded by sarcomere and metabolic defects. (A–C) Analysis of heart rate variability in larval zebrafish using high-speed imaging. Bright-field images of 120-hpf pitx2c+/+ (A) and pitx2c−/− (B) hearts. Representative kymographs of sinus rhythm (A′) and arrhythmia (B′). White dashed lines mark each cardiac cycle; blue arrows denote the duration between consecutive contractions. Box-and-whisker plot of the RMSSD between consecutive cardiac cycles (C). Each data point represents the average interval calculated from 8 to 10 cardiac cycles of individual larvae. Increased variation is observed in pitx2c−/− larvae compared with pitx2c+/+ siblings. Error bars correspond to SEM from at least 2 independent experiments. *P < 0.05 by unpaired t test. (D and E) Confocal images of cardiac sarcomeres in 50-hpf Tg(myl7:LA-EGFP); pitx2c+/+ (D) and pitx2c−/− (E) siblings. (F–I) TEM images of 56-hpf hearts (n = 3); higher magnification of the region outlined in red is shown in F’ and H’. Embryonic sarcomeres appear wavy and less organized in pitx2c−/− compared with pitx2c+/+. (J–L) Redox-state analysis of 120-hpf pitx2c+/+ and pitx2c−/− atria expressing the Tg(myl7:MITO-roGFP2-Orp1) ROS biosensor. Calibration bar shows the relative 405-/488-nm ratio. (L) Quantification of 405-/488-nm ratios in control conditions or following a 1-h HS at 37 °C prior to imaging. Following HS, increased ROS levels were detected in all 3 genotypes; a stronger increase was observed in pitx2c−/− hearts, suggesting increased levels of oxidative stress compared with pitx2c+/+ siblings (L). Error bars correspond to SEM from at least 2 independent experiments. *P < 0.05 by unpaired t test.
Developmental Changes in Sarcomeric and Metabolic Gene Expression Precede the Onset of Cardiac Arrhythmia.
To explore molecular mechanisms prior to the onset of cardiac arrhythmia, we identified Pitx2c-dependent transcriptional changes by RNA sequencing (RNA-seq) of hearts isolated from 56-hpf pitx2c+/+ and pitx2c−/− embryos. We selected this stage because pitx2c mRNA levels are higher than at 7 days postfertilization (27). In parallel, we profiled the transcriptome of hearts from a line in which pitx2c is overexpressed specifically in CMs: Tg(myl7:pitx2c-P2A-H2BGFP) (hereafter referred to as pitx2cGOF) (SI Appendix, Fig. S4). Overexpression of pitx2c in CMs resulted in a dramatic increase in atrial CM number by 48 hpf; accordingly, this overgrowth led to defective cardiac morphogenesis and lethality at early larval stages (SI Appendix, Fig. S4D).
We identified 115 differentially expressed genes comparing pitx2c+/+ with pitx2c−/− hearts and 393 differentially expressed genes comparing pitx2c+/+ with pitx2cGOF hearts (SI Appendix, Fig. S5A). Next, we performed pathway analyses based on gene expression differences comparing pitx2c+/+ with pitx2c−/− or pitx2cGOF hearts. Gene set enrichment analysis showed significant enrichment of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for carbon metabolism, oxidative phosphorylation, and cardiac muscle contraction in both pitx2c−/− (SI Appendix, Fig. S5B) and pitx2cGOF hearts (SI Appendix, Fig. S5C). Comparing these genes with a previously published Pitx2 chromatin immunoprecipitation sequencing (ChIP-seq) dataset (18) indicates that a proportion of the orthologous mouse genes have Pitx2 binding sites and thus, that at least some of the differentially expressed zebrafish genes might be direct Pitx2c targets (SI Appendix, Fig. S5D).
To validate the RNA-seq data, we performed qPCR on dissected hearts from 56-hpf pitx2c+/+, pitx2c−/−, and pitx2cGOF embryos (SI Appendix, Fig. S6). Known PITX2 target genes hcn4 and jph2 (8) were significantly up-regulated in pitx2c−/− embryonic hearts. We also measured mRNA levels of selected genes encoding metabolic regulators and found significant down-regulation of ppargc1a, which is involved in oxidative metabolism (SI Appendix, Fig. S6). Similar to what has been reported in mouse (18), we observed up-regulation of the antioxidant gene sod1 in zebrafish pitx2cGOF hearts. Furthermore, several genes involved in the cytochrome oxidase complex or mitochondrial Complexes I and III were down-regulated in pitx2c−/− and pitx2cGOF hearts. These data suggest that Pitx2c controls a set of genes that regulate the redox status of atrial CMs.
Next, we investigated whether the changes observed in mRNA levels in embryonic hearts were accompanied by sarcomere and metabolic defects. To examine sarcomere phenotypes, we analyzed 50-hpf pitx2c−/− embryos in the Tg(myl7:LA-GFP) background (Fig. 4 D and E). Subtle defects in sarcomere organization were visible by 50 hpf, prior to the stage when arrhythmia was observed. While sarcomeres in the pitx2c+/+ hearts were clearly organized perpendicular to the direction of flow (Fig. 4D), sarcomeres in pitx2c−/− hearts appeared wavy and randomly positioned, with discontinuity across adjacent CMs (Fig. 4E). These observations were also confirmed by TEM analysis (Fig. 4 F–I).
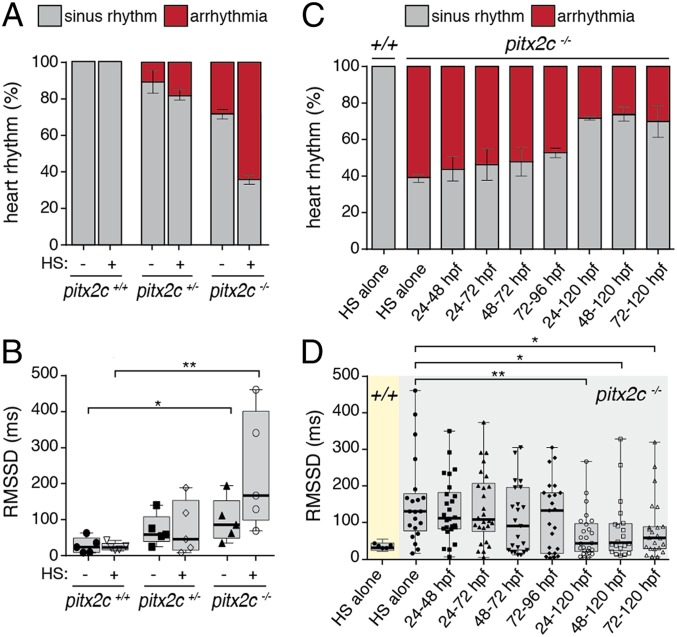
To evaluate cardiac metabolism in larvae, we analyzed the mitochondrial redox state of 120-hpf pitx2c−/− hearts using a genetically encoded roGFP2 fluorescent probe (28) (Fig. 4 J–L). Quantification of the 405-/488-nm ratios from pitx2c+/+, pitx2c+/−, and pitx2c−/− hearts at 120 hpf were not significantly different between the genotypes (Fig. 4L). To test the effect of environmental stress, we delivered a 1-h heat shock (HS) at 37 °C to 120-hpf larvae just prior to imaging. HS induced significantly higher ROS levels in pitx2c−/− hearts (1.42 ± 0.06) compared with pitx2c+/+ siblings (1.18 ± 0.04, P = 0.008), suggesting higher levels of oxidative stress in the absence of Pitx2 function (Fig. 4J). Given that the induction of ROS levels on HS was higher in pitx2c−/− larvae, we wondered whether such environmental stress could exacerbate the cardiac arrhythmia phenotype. Indeed, HS significantly increased the proportion of pitx2c−/− arrhythmic larvae (from ∼25 to ∼60%) (Fig. 5A) and RMSSD intervals (Fig. 5B).
Fig. 5.
Antioxidant treatment reduces the incidence of cardiac arrhythmia in pitx2c−/− larvae. (A and B) Effect of HS on the development of cardiac arrhythmia in 120-hpf larvae. HS (1 h at 37 °C) was performed ∼3 h prior to imaging. Proportion of larvae with sinus rhythm (gray) or arrhythmia (red; A). Box-and-whisker plots of RMSSD values in control conditions or following HS (B). Error bars indicate SEM from at least 2 independent experiments; n > 10 larvae per condition. (C and D) Treatment of larvae with 100 µM NAC at the indicated time points prior to HS and analysis of cardiac rhythm. Proportion of larvae with sinus rhythm (gray bars) or arrhythmia (red bars) following NAC treatment (C). Box-and-whisker plots of RMSSD values from pitx2c+/+ (yellow) and pitx2c−/− (gray) in control conditions or following NAC treatment (D). Each data point represents the average interval calculated from 8 to 10 cardiac cycles of individual larvae. Treatment of pitx2c−/− larvae starting from 24, 48, or 72 to 120 hpf significantly reduced the number of arrhythmic hearts compared with HS alone (D). Error bars indicate SEM from at least 2 independent experiments. n > 20 pitx2c−/− animals per condition. *P < 0.05 by unpaired t test; **P < 0.01 by unpaired t test.
Metabolic Stress Is Required, but Not Sufficient, to Drive Cardiac Arrhythmia in Zebrafish Larvae.
Transcriptional profiling and functional data suggest that both sarcomere and metabolic abnormalities are present in pitx2c−/− larvae prior to the onset of cardiac arrhythmia. Therefore, we next queried whether the contribution of these two factors could be separated. To test this possibility, embryos from pitx2c+/− incrosses were treated with 100 μM NAC, a potent ROS scavenger. Different treatment protocols were used to determine a precise temporal window of efficacy between 24 and 120 hpf. While HS did not induce cardiac arrhythmia in pitx2c+/+ siblings, 60% of pitx2c−/− larvae were arrhythmic (Fig. 5C, HS alone), with significant beat-to-beat variability (157.2 ± 25.9 ms, P = 0.017) (Fig. 5D, HS alone). Continuous treatment with NAC from 24 to 120 hpf significantly decreased the proportion of pitx2c−/− larvae with cardiac arrhythmia to ∼25% (Fig. 5C) and reduced heart rate variability to 64.4 ± 12.3 ms (P = 0.0014) (Fig. 5D). These data suggest that increased ROS levels are an important driver of cardiac arrhythmia.
To investigate whether metabolic stress alone could trigger arrhythmia in the absence of sarcomere defects, we analyzed 2 models of metabolic insufficiency: Mia40a, an oxidoreductase essential for mitochondrial protein biogenesis (29, 30), and Ppargc1a, a master regulator of oxidative metabolism (31). In both models, we failed to observe a significant number of hearts that were arrhythmic, although HS did lead to more variability in cardiac intervals in both mia40a−/− (45.00 ± 10.64 and 56.08 ± 58.40 ms in control and HS, respectively) and ppargc1a−/− (22.60 ± 3.47 and 52.08 ± 38.52 ms in control and HS, respectively) compared with heterozygous and wild-type siblings (SI Appendix, Fig. S7). Taken together, these data suggest that metabolic stress is necessary, but not sufficient, for the development of arrhythmias.
Discussion
Zebrafish offer unique advantages to study adult cardiac phenotypes. Compared with rodents, many aspects of zebrafish cardiac electrophysiology are more closely aligned with those of humans, including heart rate, developmental physiology, and cardiac action potential characteristics (32, 33). Indeed, modeling human arrhythmia in zebrafish has provided significant insight into mechanisms of conduction defects (33–36). Additionally, most dilated cardiomyopathy genes in humans have a corresponding ortholog in zebrafish (37), and zebrafish mutants in these genes reveal phenotypes resembling human cardiomyopathies (38, 39). Thus, from genetic, developmental, and electrophysiological perspectives, the zebrafish model is highly complementary to established in vivo models and may offer particular strengths over some of the existing models.
In this study, we describe the phenotypes observed in larval and adult zebrafish hearts following loss of Pitx2c function and propose that the pitx2c mutant can serve as a model of cardiac arrhythmia. We find atrial conduction defects, structural remodeling, and altered cardiac metabolism resembling the phenotypes observed in human AF patients. In comparison with rodent models of AF, pitx2c−/− zebrafish display an arrhythmia phenotype without pacing, thus representing an animal model of spontaneous arrhythmia. Furthermore, we detected cardiac phenotypes in a subset of adult pitx2c+/− zebrafish, similar to what is observed in PITX2+/− humans and mice (5).
Traditionally, AF has been primarily considered an electrophysiological disease. Indeed, Pitx2 has been shown to directly regulate targets, including ion channel genes, that modulate electrical conduction properties. Our RNA-seq analysis of both gain- and loss-of-function embryonic hearts revealed dysregulation of some of these targets, including ryr2, cacna1g, jph2, and kcnq1 as well as atp2a1 and atp2a2. These data indicate that Pitx2 regulates ion channel gene expression during cardiac development in zebrafish as has been shown in mouse (8, 9). We hypothesize that altered expression of ion channel genes (e.g., hcn4) in conjunction with sarcomere and metabolic defects in embryonic and adult hearts all contribute to the observed cardiac phenotypes.
In recent years, several reports have expanded our understanding of AF as a complex multifactorial disease. These studies suggest that mutations in genes that compromise sarcomere structure can also lead to AF phenotypes (19–21, 40). Our analyses of pitx2c−/− adult atria revealed significantly compromised cardiac tissue integrity by histology and TEM analysis as well as cardiac dysfunction by echocardiography. Additionally, we detected alterations in sarcomere organization in embryos long before extensive remodeling and electrophysiological defects are observed in adults. RNA-seq profiling of embryonic hearts also identified many differentially expressed genes associated with cardiac contraction. Many of these features are also observed in models of hypertrophic cardiomyopathy (41), and we, therefore, hypothesize that an underlying atrial cardiomyopathy precedes the development of arrhythmia, fibrosis, and other hallmarks of AF. These zebrafish data are in further support of the hypothesis that atrial cardiomyopathy predisposes the human heart to AF (19–21, 40).
Pitx2 activates a set of stress-response genes during skeletal muscle development (42) as well as during cardiac regeneration (18). Furthermore, Pitx2 is important for the maintenance of mitochondrial structure and function (25) and regulates the expression of mitochondrial genes in the mouse heart (18, 25). We found that some of these genes are similarly dysregulated in zebrafish embryonic hearts and observed increased ROS levels in pitx2c−/− larval hearts. As Pitx2c expression is enriched in the atria of zebrafish and mice, we hypothesize that it modulates the expression of metabolic genes in this chamber. We also observed in pitx2c−/− atria increased mtDNA content without a significant impact on mitochondrial respiratory capacity; this presumed increase in the number of mitochondria without a concomitant increase in respiration suggests a defect in mitochondrial function.
We observed that continuous NAC treatment reduced the proportion of arrhythmic pitx2c−/− larvae, indicating that increased ROS levels contribute to the development of the cardiac arrhythmia phenotype. However, we did not observe an arrhythmogenic phenotype in mia40a and ppargc1a models of metabolic insufficiency, suggesting that arrhythmias do not arise solely due to metabolic alterations. Altogether, these data indicate that metabolic stress is required, but not sufficient, for the development of cardiac arrhythmia in larvae. As HS treatment increased ROS levels more significantly in pitx2c−/− larval hearts compared with pitx2c+/+, we hypothesize that pitx2c mutants are more sensitive than wild types to environmental stress, perhaps in part by a reduction in ROS scavengers. Therefore, increased ROS levels in conjunction with cardiac sarcomere defects contribute to the development of a cardiac arrhythmia phenotype in zebrafish larvae. It is also important to consider that several sarcomeric proteins are ROS sensitive. In many cases, redox modifications resulting in oxidation or nitrosylation can disrupt the function of such proteins and contribute to contractile dysfunction (43–45). This aspect of cross-talk between sarcomere and metabolic defects may also be highly relevant in the pitx2c model of cardiac arrhythmia.
Altogether, our data support the hypothesis that sarcomere defects in conjunction with metabolic stress are required for the development of electrical dysfunction and other hallmarks of AF. We propose a model whereby sarcomere and metabolic defects are present in the pitx2c−/− embryonic heart, and they act as arrhythmogenic substrates already at these early stages of development (SI Appendix, Fig. S8). With age and continued cardiac stress, phenotypes such as atrial conduction defects and structural remodeling are observed. We hypothesize that atrial energetics attempt to compensate by increased mitochondrial respiration, which may consequently lead to oxidative stress. At late adult stages, many of the phenotypes observed in pitx2c−/− hearts are suggestive of heart failure, recapitulating the proposed end point diagnosis of some untreated AF patients. In summary, we propose that the pathogenesis of arrhythmia and AF-like phenotypes can be driven by early sarcomere and metabolic changes that lead to atrial myopathy and altered atrial cardiac energetics.
Materials and Methods
Detailed methods can be found in SI Appendix.
Zebrafish Husbandry.
Zebrafish husbandry was performed under standard conditions in accordance with the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes. Procedures involving animals were approved by the veterinary department of the Regional Board of Darmstadt and the Danish Animal Experimental board (personal project license 2017-15-0201-01305).
ECG Analysis.
Adult zebrafish were anesthetized in 0.03% Tricaine/MS-222 (Sigma-Aldrich) in fish tank water, and the zebrafish were placed dorsal side down in a damp sponge. ECG traces were analyzed beat per beat using a minimum of 30 consecutive beats.
RNA-seq.
Detailed methods can be found in SI Appendix. RNA-seq datasets have been deposited to the Gene Expression Omnibus repository under accession number GSE128511.
Data Analysis.
Images were analyzed using ImageJ/FIJI and Zen software (Zeiss). Statistical analyses were performed using Prism (GraphPad).
Data Availability Statement.
All data discussed in the paper are available to readers in the Gene Expression Omnibus repository under accession number GSE128511.
Supplementary Material
Acknowledgments
We thank M. Santoro and E. Panieri for the Tg(myl7:MITO-roGFP2-Sce.Hyr1)uto54Tg line and imaging advice; J. Martin and M. Zhang for sharing the Pitx2 datasets; H. Schultheis and M. Looso for bioinformatics support; B. Berge for TEM assistance; S. Allanki for sharing reagents; S. Howard, B. Grohmann, M. Ploch, P. Neeb, and C. Kremser for technical support; and members of the laboratory of D.Y.R.S. for fruitful discussions. This work was supported by funds from the Deutsches Zentrum für Herz-Kreislauf-Forschung (German Centre for Cardiovascular Research, to M.M.C.), the Novo Nordisk Foundation (to M.S.O. and P.R.L.), the John and Birthe Meyer Foundation (to M.S.O. and P.R.L.), the Deutsche Forschungsgemeinschaft (project no. 394046768 SFB1366/project A4) (to R.M.-J. and D.Y.R.S.), and the Max Planck Society and Leducq Foundation (to D.Y.R.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. C.A.M. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE128511).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913905116/-/DCSupplemental.
References
- 1.Gudbjartsson D. F., et al. , Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature 448, 353–357 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Ellinor P. T., et al. , Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat. Genet. 44, 670–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lubitz S. A., et al. , Novel genetic markers associate with atrial fibrillation risk in Europeans and Japanese. J. Am. Coll. Cardiol. 63, 1200–1210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roselli C., et al. , Multi-ethnic genome-wide association study for atrial fibrillation. Nat. Genet. 50, 1225–1233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J., et al. , Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc. Natl. Acad. Sci. U.S.A. 107, 9753–9758 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirchhof P., et al. , PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ. Cardiovasc. Genet. 4, 123–133 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Ammirabile G., et al. , Pitx2 confers left morphological, molecular, and functional identity to the sinus venosus myocardium. Cardiovasc. Res. 93, 291–301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao Y., et al. , Pitx2, an atrial fibrillation predisposition gene, directly regulates ion transport and intercalated disc genes. Circ. Cardiovasc. Genet. 7, 23–32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadadur R. D., et al. , Pitx2 modulates a Tbx5-dependent gene regulatory network to maintain atrial rhythm. Sci. Transl. Med. 8, 354ra115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lozano-Velasco E., et al. , Pitx2 impairs calcium handling in a dose-dependent manner by modulating Wnt signalling. Cardiovasc. Res. 109, 55–66 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Aguirre L. A., et al. , Long-range regulatory interactions at the 4q25 atrial fibrillation risk locus involve PITX2c and ENPEP. BMC Biol. 13, 26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chinchilla A., et al. , PITX2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ. Cardiovasc. Genet. 4, 269–279 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Mora C., et al. , Generation of induced pluripotent stem cells (iPSC) from an atrial fibrillation patient carrying a PITX2 p.M200V mutation. Stem Cell Res. 24, 8–11 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Mechakra A., et al. , A novel PITX2c gain-of-function mutation, p.Met207Val, in patients with familial atrial fibrillation. Am. J. Cardiol. 123, 787–793 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Ryan A. K., et al. , Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature 394, 545–551 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Piedra M. E., Icardo J. M., Albajar M., Rodriguez-Rey J. C., Ros M. A., Pitx2 participates in the late phase of the pathway controlling left-right asymmetry. Cell 94, 319–324 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Logan M., Pagan-Westphal S. M., Smith D. M., Paganessi L., Tabin C. J., The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell 94, 307–317 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Tao G., et al. , Pitx2 promotes heart repair by activating the antioxidant response after cardiac injury. Nature 534, 119–123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orr N., et al. , A mutation in the atrial-specific myosin light chain gene (MYL4) causes familial atrial fibrillation. Nat. Commun. 7, 11303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudbjartsson D. F., et al. , A frameshift deletion in the sarcomere gene MYL4 causes early-onset familial atrial fibrillation. Eur. Heart J. 38, 27–34 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Ahlberg G., et al. , Rare truncating variants in the sarcomeric protein titin associate with familial and early-onset atrial fibrillation. Nat. Commun. 9, 4316 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins M. M., et al. , Pitx2c orchestrates embryonic axis extension via mesendodermal cell migration. eLife 7, e34880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korantzopoulos P., Kolettis T. M., Galaris D., Goudevenos J. A., The role of oxidative stress in the pathogenesis and perpetuation of atrial fibrillation. Int. J. Cardiol. 115, 135–143 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Huang C. X., Liu Y., Xia W. F., Tang Y. H., Huang H., Oxidative stress: A possible pathogenesis of atrial fibrillation. Med. Hypotheses 72, 466–467 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Li L., et al. , Pitx2 maintains mitochondrial function during regeneration to prevent myocardial fat deposition. Development 145, dev168609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaffer F., Ginsberg J. P., An overview of heart rate variability metrics and norms. Front. Public Health 5, 258 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunawan F., et al. , Focal adhesions are essential to drive zebrafish heart valve morphogenesis. J. Cell Biol. 218, 1039–1054 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panieri E., Millia C., Santoro M. M., Real-time quantification of subcellular H2O2 and glutathione redox potential in living cardiovascular tissues. Free Radic. Biol. Med. 109, 189–200 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Sokol A. M., et al. , Loss of the Mia40a oxidoreductase leads to hepato-pancreatic insufficiency in zebrafish. PLoS Genet. 14, e1007743 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chacinska A., et al. , Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 23, 3735–3746 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehman J. J., et al. , Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Invest. 106, 847–856 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravens U., Ionic basis of cardiac electrophysiology in zebrafish compared to human hearts. Prog. Biophys. Mol. Biol. 138, 38–44 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Milan D. J., Macrae C. A., Zebrafish genetic models for arrhythmia. Prog. Biophys. Mol. Biol. 98, 301–308 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chi N. C., et al. , Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 6, e109 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ebert A. M., et al. , Calcium extrusion is critical for cardiac morphogenesis and rhythm in embryonic zebrafish hearts. Proc. Natl. Acad. Sci. U.S.A. 102, 17705–17710 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnaout R., et al. , Zebrafish model for human long QT syndrome. Proc. Natl. Acad. Sci. U.S.A. 104, 11316–11321 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shih Y. H., et al. , Cardiac transcriptome and dilated cardiomyopathy genes in zebrafish. Circ. Cardiovasc. Genet. 8, 261–269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bakkers J., Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc. Res. 91, 279–288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gut P., Reischauer S., Stainier D. Y. R., Arnaout R., Little fish, big data: Zebrafish as a model for cardiovascular and metabolic disease. Physiol. Rev. 97, 889–938 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi S. H., et al. ; DiscovEHR study and the NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium , Association between titin loss-of-function variants and early-onset atrial fibrillation. J. Am. Med. Assoc. 320, 2354–2364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dvornikov A. V., de Tombe P. P., Xu X., Phenotyping cardiomyopathy in adult zebrafish. Prog. Biophys. Mol. Biol. 138, 116–125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.L’honoré A., et al. , Redox regulation by Pitx2 and Pitx3 is critical for fetal myogenesis. Dev. Cell 29, 392–405 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Janué A., Odena M. A., Oliveira E., Olivé M., Ferrer I., Desmin is oxidized and nitrated in affected muscles in myotilinopathies and desminopathies. J. Neuropathol. Exp. Neurol. 66, 711–723 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Grützner A., et al. , Modulation of titin-based stiffness by disulfide bonding in the cardiac titin N2-B unique sequence. Biophys. J. 97, 825–834 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steinberg S. F., Oxidative stress and sarcomeric proteins. Circ. Res. 112, 393–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the paper are available to readers in the Gene Expression Omnibus repository under accession number GSE128511.