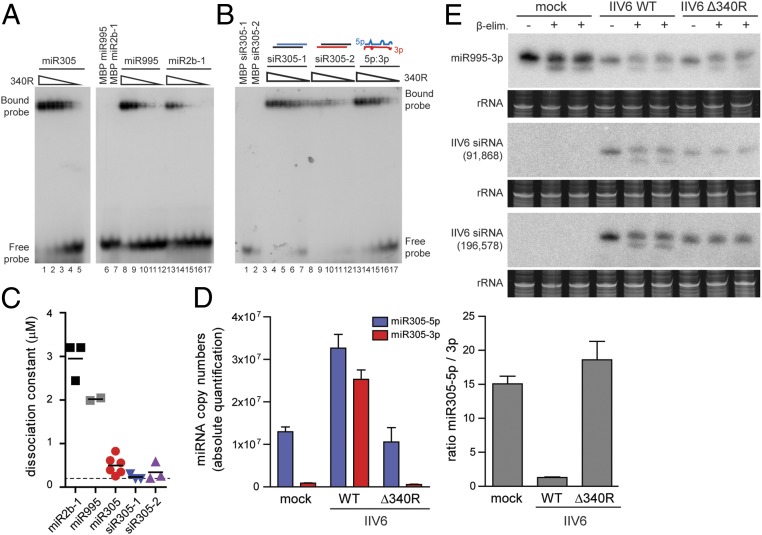
Fig. 4.
340R efficiently binds miR-305 and siRNA duplexes. (A) EMSA of 5p:3p duplexes of miR-305, miR-995, and miR-2b-1 with recombinant 340R. (B) EMSA of the miR-305 5p:3p duplex (lanes 13 to 17) and miR-305–derived siRNAs with 340R. The siRNAs were designed by annealing miR-305–5p or miR-305–3p with perfectly complementary RNA to generate siRNAs with 2-nt 3′ overhangs (siR305-1 and siR305-2, respectively; lanes 3 to 7 and lanes 8 to 12). Two-fold dilutions of recombinant protein were tested, starting from a concentration of 3.2 μM. Incubations in buffer only (−) and MBP were included as controls, as indicated. (C) Dissociation constants of 340R for the indicated miRNA and siRNA duplexes. Each symbol represents an independent experiment. Horizontal lines indicate means. The dashed line is the lowest concentration tested. (D) Absolute quantification of miR-305–5p and miR-305–3p by stemloop-qPCR in mock infected cells and in IIV6 WT and Δ340R-infected S2 cells at 72 hpi. For each miRNA, a standard curve of synthetic RNA was used for quantification. Expression of the miRNAs was normalized to expression of the U6 small nuclear RNA. (E) Northern blot analysis of miR-995–3p and 2 IIV6-derived siRNAs (positions in the genome indicated) in total RNA of Drosophila S2 cells infected with IIV6 WT or Δ340R at 3 dpi. Where indicated, the samples were subjected to β-elimination (+) or mock treated (−), with 2 technical replicates on the same RNA for the β-elimination reaction. Ethidium-bromide–stained rRNA was used as a loading control.