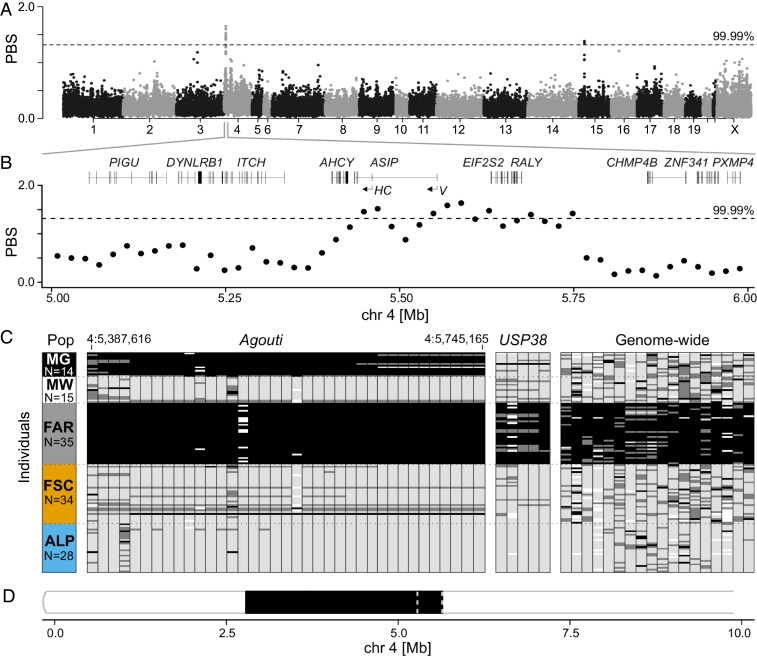
Fig. 2.
The candidate association region for winter-white/winter-gray coat color polymorphism in mountain hares. (A) Genome scan of PBS for Faroese hares in 20-kb nonoverlapping windows. (B) Zoomed-in view of PBS in the chromosome 4 candidate region and gene structure (noncoding exons of Agouti are marked as HC, hair-cycle specific isoform; V, ventral isoform). The dashed line represents the 0.01% genome-wide cutoff. (C) Genotypes at 60 loci spanning the Agouti (chromosome 4) and USP38 (chromosome 15) candidate regions, and other windows of high PBS along the genome (SI Appendix, Tables S6 and S7 and Fig. S5). Rows depict specimens and columns indicate genotyped loci (coordinates of first and last SNPs in the Agouti region are indicated). Black, homozygous for the Faroese variant; light gray, homozygous for the alternative variant; dark gray, heterozygous; white, missing data; ALP, Alps; FAR, Faroe Islands; FSC, Fennoscandia; MG, winter-gray museum specimens; MW, winter-white museum specimens. (D) Selective sweep detected by Pool-hmm in the Faroese hares on the first 10 Mb of chromosome 4; dashed lines delimit the association region (5.40–5.76 Mb).