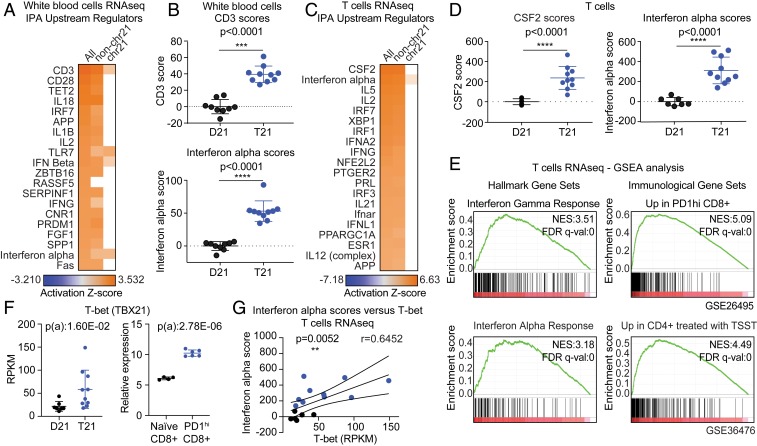
Fig. 6.
Transcriptome analysis reveals gene expression signatures indicative of T cell activation in adults with trisomy 21. RNAseq analysis was performed using bulk WBCs (n = 9 D21; n = 10 T21). (A) Heatmap displaying the results of examining the RNAseq data using the Upstream Regulator tool of the IPA software. Factors predicted to drive the gene expression changes observed are labeled to the left of the heatmap, with CD3 on top. From left to right, the analysis was performed using all DEGs, DEGs not encoded on chr21 (non-chr21), and DEGs encoded on chr21. (B) Dot and whisker plots showing (Top) CD3 activation scores and (Bottom) IFN alpha scores derived from WBC samples with and without T21. (C) Upstream regulator analysis as in A using RNAseq data derived from bulk T cells (n = 7 D21; n = 10 T21). (D) Dot and whisker plots of (Left) CSF2 scores and (Right) IFN alpha scores derived from T cells with and without T21. (E) Enrichment plots from GSEA of T cell RNAseq data, using (Left) the Hallmarks gene sets or (Right) Immunological Signature gene sets. Normalized Enrichment Score (NES) and FDR-corrected q values are shown. (F) Dot and whisker plots for T-bet mRNA levels from (Left) T cell RNAseq data and (Right) the GSE26495 dataset. Benjamini−Hochberg adjusted P values were calculated using DESeq2 and GEO2R (ebayes), respectively. (G) Pearson correlation analysis for IFN alpha scores versus T-bet mRNA expression in T cells. Data in B, D, and F are shown as mean ± SD with significance determined by unpaired t test; data in G are shown as correlation with significance determined by Pearson’s correlation. **P < 0.01; ***P < 0.001 ****P < 0.0001.