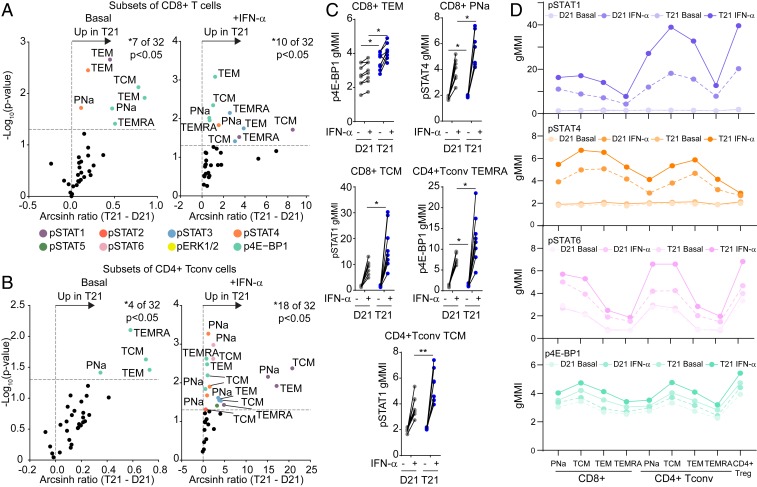
Fig. 7.
T cells of people with trisomy 21 are hypersensitive to IFN-α stimulation. Whole blood was incubated directly ex vivo for 30 min with (+IFN-α) or without (basal) IFNα-2A, before being processed for mass cytometry analysis of 8 different phosphoepitopes in 9 different T cell subsets (see SI Appendix, Fig. S8A for gating strategy). (A and B) Volcano plots showing fold changes by karyotype (T21 versus D21) either (Left) under basal conditions or (Right) after stimulation with IFNα-2A on subsets of (A) CD8+ T cells and (B) CD4+ Tconv cells. Fold change was calculated as the arcsinh ratio of samples from people with T21 minus that of samples of typical people (D21). All data represent one Helios run containing a total of n = 8 biologically independent replicates per group. Vertical dashed line represents the no-change midline. Horizontal dashed line represents P value of 0.05 as calculated by Student t test. (C) Dot plots displaying geometric mean metal intensity (gMMI) for the indicated epitopes and cell types, with lines connecting the basal and IFN-α−stimulated values for each sample. (D) Scatter plots showing the gMMI of pSTAT1, pSTAT4, pSTAT6, and p4E-BP1 among the indicated T cell subsets in people with and without T21 before and after stimulation with IFNα-2A. Data in C are shown as mean ± SEM with significance determined by 2-way ANOVA with Sidak’s posttest. *P < 0.05; **P < 0.01.