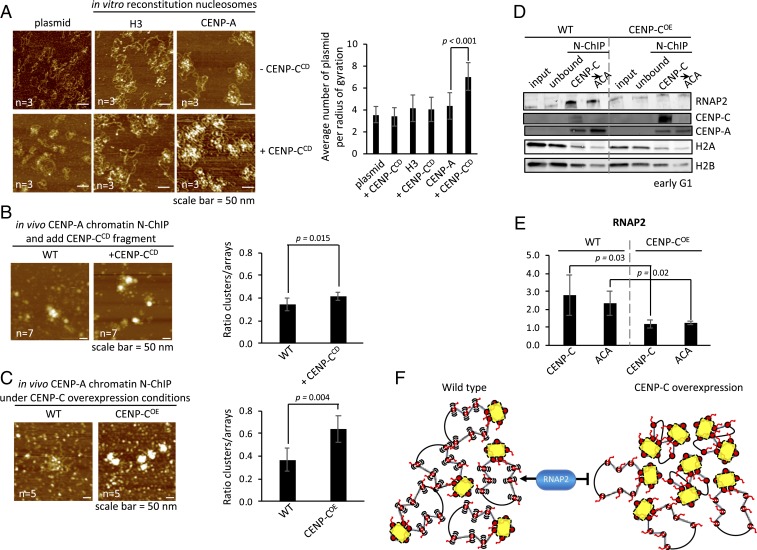
Fig. 4.
CENP-C overexpression compacts CENP-A chromatin, making it inaccessible to RNAP2. (A) Quantitative assessment of in vitro reconstituted chromatin showed that only CENP-A chromatin clustered in the presence of CENP-CCD fragment, whereas H3 chromatin or naked plasmids did not in the presence of CENP-CCD. Plasmid clustering was measured by counting the number of plasmids in a radius of gyration (r = 0.25 µm). (B) To determine if the CENP-CCD fragment used in the in vitro experiments could induce CENP-A chromatin compaction, we added CENP-CCD for 30 min to isolated free CENP-A chromatin from HeLa cells. Nucleosome arrays can be identified as either bead-on-a-string or large compacted clusters where DNA strands can be seen entering/exiting. Compacted chromatin was scored over the total number of nucleosome arrays. (C) Similar analysis was performed on unbound CENP-A chromatin extracted from cells that either did (CENP-COE) or did not (WT) overexpress CENP-C. (D) Centromeres are expressed during early G1. Therefore, we synchronized HeLa cells to early G1 and extracted kinetochore-bound (first CENP-C N-ChIP) and unbound CENP-A chromatin (second ACA N-ChIP of unbound fraction; see Methods for details). By Western blot we probed for RNAP2, CENP-C, CENP-A, H2A, and H2B. (E) Quantification of RNAP2 levels were determined by LiCor’s software. The bar graphs represent 3 independent experiments. (F) Working model of CENP-C (yellow) overexpression inducing CENP-A chromatin (red) cross-array clustering thereby reducing access to RNAP2 (blue).