Fig. 2.
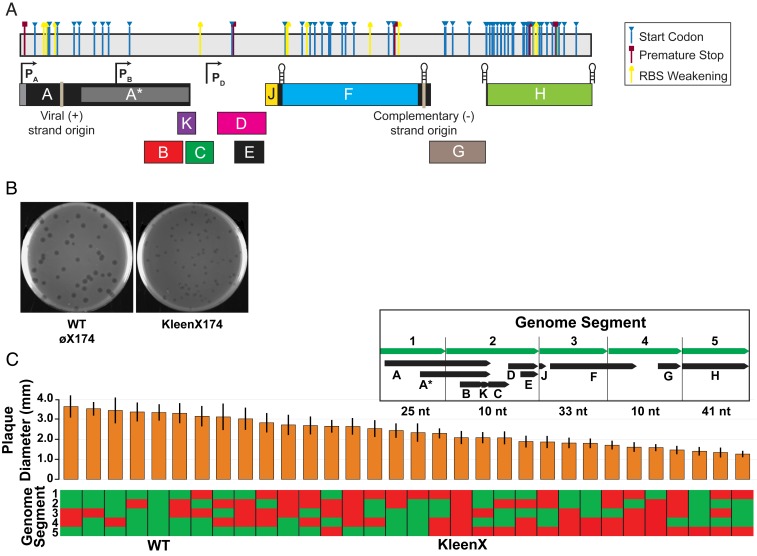
A “clean” øX174 genome in which 71 cryptic ORFs are simultaneously disrupted is viable but has reduced fitness. (A) Linear depiction of kleenX174 genome showing the locations and modes of cryptic ORF disruption; see Dataset S1 for detailed information. (B) Plaques of wild-type and kleenX174 phage. 85 mm diameter plates. (C) Plaque diameter of wild-type/kleenX174 chimeras, arranged from largest average plaque size to smallest. Vertical bars represent 1 SD from n = 50 plaque measurements. Each chimeric phage consists of 5 genome segments chosen from a mixture of wild-type genome segment (green) and kleenX174 modified genome segments (red). (Inset) The boundaries of the 5 genome segments, protein-coding ORFs found in each segment, and total number of nucleotide differences between wild-type øX174 and kleenX174 genome sequences in each segment.