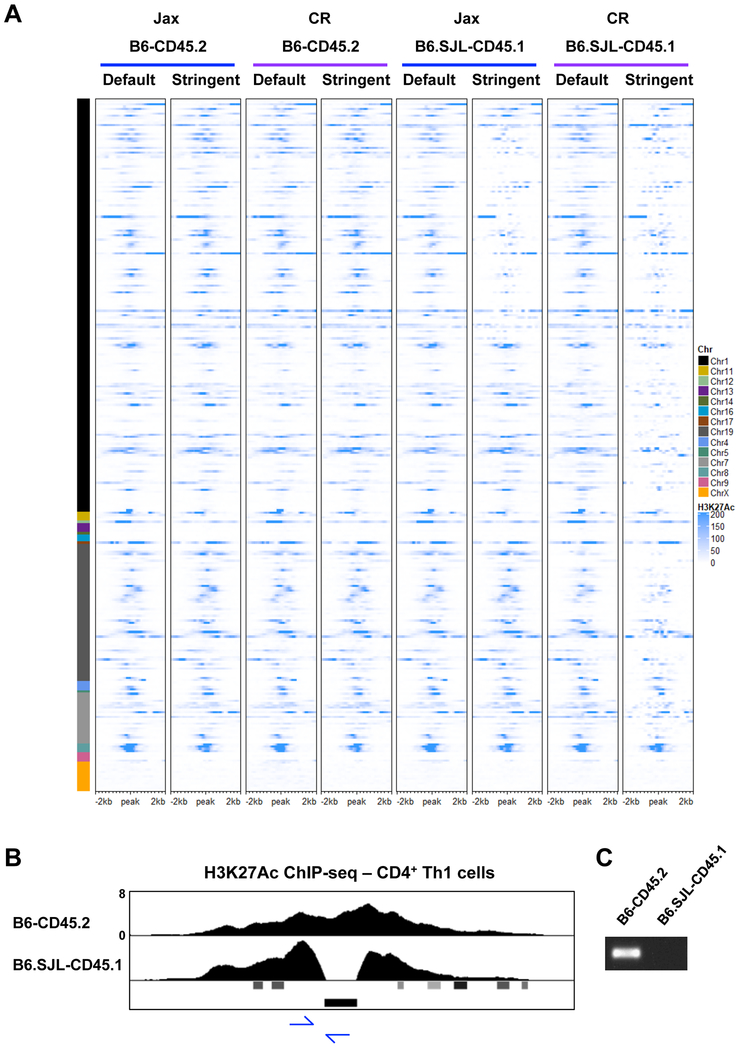
Figure 6. Genetic variation is found in H3K27Ac elements in CD4+ Th1 cells.
(A) Heatmap of H3K27Ac ChIP-seq datasets comparing default and stringent alignment parameters for CD4+ Th1 cells isolated from B6-CD45.2 and B6.SJL-CD45.1 mice from Jax or CR. See results and methods for heatmap peak selection. (B) UCSC genome browser display of H3K27Ac ChIP-seq tracks for CD4+ Th1 cells isolated from B6-CD45.2 or B6.SJL-CD45.1 mice from Jax. Viral LTR annotation is shown below the tracks. Primer locations for PCR in (C) are shown with blue arrows. (C) PCR analysis examining DNA from B6-CD45.2 or B6.SJL-CD45.1 mice from Jax. (A-C) Data are representative of at least 2 independent biological replicates. See also Fig. S5.