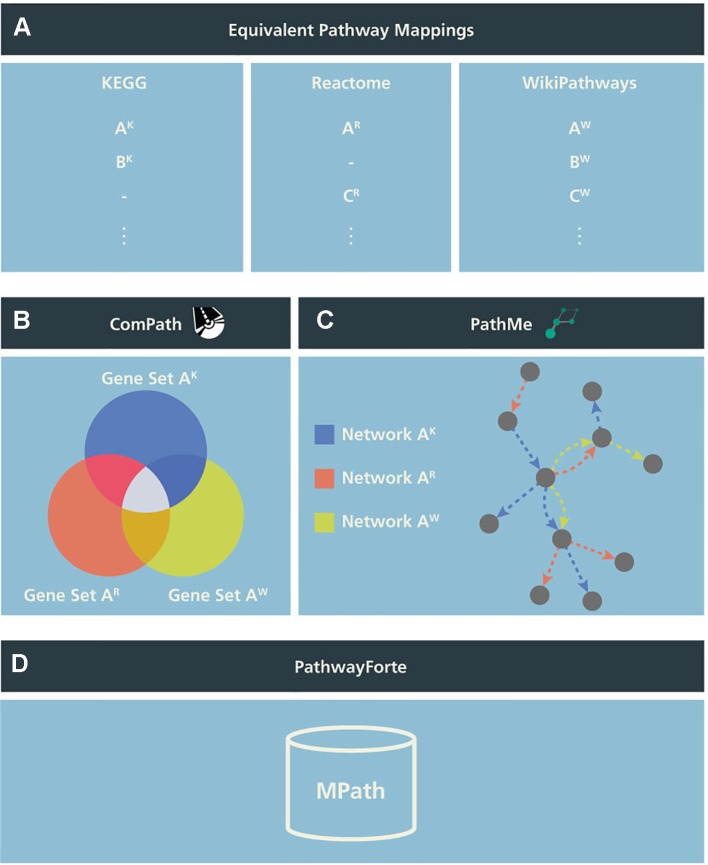
Figure 1.
Schema illustrating the generation of MPath. The curated pathway mapping catalog is depicted in (A), which links equivalent pathways from different resources. Pathways that are shared across two resources are referred to as pathway analogs (i.e., Pathway A in Reactome and Pathway A′ in KEGG) and pathways that are shared across all three resources are referred to as “super pathways” (i.e., Pathway A in KEGG, Pathway A′ in Reactome, and Pathway A″ in WikiPathways). (B) Using these mappings, gene sets of equivalent pathways from different resources can be combined, ensuring key molecular players from the different resources are included. (C) Similarly, network representations of the pathways can be overlaid to generate more comprehensive pathways. (D) Finally, both the combined gene sets and networks representations are included in MPath. Note that pathways that are exclusive to a single database are included in MPath unchanged.