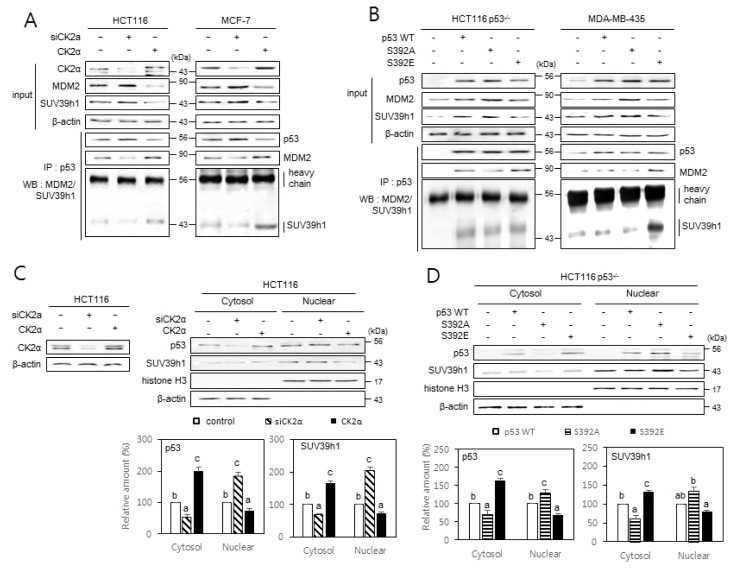
Fig. 5. The effect of p53 S392 phosphorylation on the interactions between p53, MDM2, and SUV39h1, and the nuclear localization of SUV39h1.
Cells were treated with CK2α siRNA or pcDNA3.1-CK2 (A and C) or with pcDNA3.1-p53 variants (B and D). (A and B) The inhibitory effect of CK2 downregulation on the interactions between p53, MDM2, and SUV39h1 (A) and the induction of p53, MDM2, and SUV39h1 interactions by p53S392E relative to wild-type (WT) p53 and p53S392A (B). Cell lysates were immunoprecipitated (IP) with anti-p53 antibodies followed by immunoblotting with anti-MDM2 and SUV39h1 antibodies. Cell lysate IP with IgG heavy chain served as a loading control. The same membranes were reprobed with anti-p53 antibodies and TrueBlot reagent, which eliminates the detection of the immunoprecipitated antibodies. Representative data from three independent experiments are shown. (C and D) The inhibitory effect of CK2 upregulation on the nuclear localization of p53 and SUV39h1 (C) and the induction of SUV39h1 nuclear localization by p53S392A relative to WT p53 and p53S392E (D). Cytoplasm and nuclei were isolated from p53-null cells overexpressing variants of p53 and were visualized by immunoblotting. β-actin (cytoplasmic marker) and histone H3 (nuclear marker) were quantified as loading controls (upper panels). The levels of CK2α protein in whole cells extracts are shown (left on the C panel). The graph shows the quantification of p53 and SUV39h1 relative to the subcellular markers (bottom panels). The values indicate mean ± SE. Bars that do not share a common letter (a, b, or c) are significantly different between groups at P < 0.05.